Coverage of endogenous genes - single cell reads
2015-02-23
Last updated: 2016-02-23
Code version: 70509eb9a08ffe0fe459efc9de23d89ec424fe99
The sequencing coverage from the UMI protocol should show a very strong 5’ bias. Do we observe this in our data? Here we explore this in a few samples using the genomation package. Specifically, we calculate the mean coverage across all the genes that passed our expression filters for two regions:
- The transcription start site (TSS) +/- 1 kb
- The transcription end site (TES) +/- 1 kb
Using the single cell reads, we observe the same pattern as with the molecules.
library("genomation")
library("Rsamtools")
library("plyr")
library("tidyr")
library("ggplot2")
theme_set(theme_bw(base_size = 14))
theme_update(panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.major.y = element_blank())Input
Input filtered read counts.
reads_filter <- read.table("../data/reads-filter.txt", header = TRUE,
stringsAsFactors = FALSE)Prepare bam files from high quality cells
To investigate the coverage, we select one high quality cell from each individual. Note that for the actual data files, the names are more succinct to keep them shorter.
quality_cells <- c("19098.1.A01", "19101.1.A02", "19239.1.A01")
names(quality_cells) <- c("NA19098.r1.A01", "NA19101.r1.A02", "NA19239.r1.A01")
stopifnot(names(quality_cells) %in% colnames(reads_filter))From the sequencing pipeline, the combined bam files for the reads are in bam-combined and have the filename structure individual.replicate.well.trim.sickle.sorted.combined.bam. These files are already sorted and indexed.
bam_reads <- paste0(quality_cells, ".trim.sickle.sorted.combined.bam")
data_dir <- "/mnt/gluster/home/jdblischak/ssd"
from_file <- file.path(data_dir, "bam-combined", bam_reads)
to_file <- file.path("../data", bam_reads)
indexed_file <- paste0(bam_reads, ".bai")
from_file_index <- file.path(data_dir, "bam-combined", indexed_file)
to_file_index <- file.path("../data", indexed_file)
for (f in 1:length(bam_reads)) {
if (!file.exists(to_file_index[f])) {
stopifnot(file.exists(from_file[f], from_file_index[f]))
file.copy(from_file[f], to_file[f])
file.copy(from_file_index[f], to_file_index[f])
}
}
stopifnot(file.exists(to_file, to_file_index))
bam <- to_file
bam[1] "../data/19098.1.A01.trim.sickle.sorted.combined.bam"
[2] "../data/19101.1.A02.trim.sickle.sorted.combined.bam"
[3] "../data/19239.1.A01.trim.sickle.sorted.combined.bam"Prepare genomic features
The genomic features are created with the script create-transcripts.R.
Input transcription start sites (TSS).
tss <- readBed("../data/tss.bed")
tss <- tss[tss$name %in% rownames(reads_filter)]Input transcription end sites (TES).
tes <- readBed("../data/tes.bed")
tes <- tes[tes$name %in% rownames(reads_filter)]Calculate coverage over genomic features
TSS
tss_sm = ScoreMatrixList(target = bam, windows = tss, type = "bam",
rpm = TRUE, strand.aware = TRUE)working on: 19098.1.A01.trim.sickle.sorted.combined.bam
Normalizing to rpm ...
working on: 19101.1.A02.trim.sickle.sorted.combined.bam
Normalizing to rpm ...
working on: 19239.1.A01.trim.sickle.sorted.combined.bam
Normalizing to rpm ...tss_smscoreMatrixlist of length:3
1. scoreMatrix with dims: 12192 2001
2. scoreMatrix with dims: 12192 2001
3. scoreMatrix with dims: 12192 2001TES
tes_sm = ScoreMatrixList(target = bam, windows = tes, type = "bam",
rpm = TRUE, strand.aware = TRUE)working on: 19098.1.A01.trim.sickle.sorted.combined.bam
Normalizing to rpm ...
working on: 19101.1.A02.trim.sickle.sorted.combined.bam
Normalizing to rpm ...
working on: 19239.1.A01.trim.sickle.sorted.combined.bam
Normalizing to rpm ...tes_smscoreMatrixlist of length:3
1. scoreMatrix with dims: 12191 2001
2. scoreMatrix with dims: 12191 2001
3. scoreMatrix with dims: 12191 2001Summarize coverage
Calculate the mean coverage per base pair for the TSS and and TES.
names(tss_sm) <- names(quality_cells)
tss_sm_df <- ldply(tss_sm, colMeans, .id = "sample_id")
colnames(tss_sm_df)[-1] <- paste0("p", 1:(ncol(tss_sm_df) - 1))
tss_sm_df$feature = "TSS"
tss_sm_df_long <- gather(tss_sm_df, key = "pos", value = "rpm", p1:p2001)names(tes_sm) <- names(quality_cells)
tes_sm_df <- ldply(tes_sm, colMeans, .id = "sample_id")
colnames(tes_sm_df)[-1] <- paste0("p", 1:(ncol(tes_sm_df) - 1))
tes_sm_df$feature = "TES"
tes_sm_df_long <- gather(tes_sm_df, key = "pos", value = "rpm", p1:p2001)Combine the two features.
features <- rbind(tss_sm_df_long, tes_sm_df_long)
# Convert base position back to integer value
features$pos <- sub("p", "", features$pos)
features$pos <- as.numeric(features$pos)
# Subtract 1001 to recalibrate as +/- 1 kb
features$pos <- features$pos - 1001
# Order factor so that TSS is displayed left of TES
features$feature <- factor(features$feature, levels = c("TSS", "TES"),
labels = c("Transcription start site",
"Transription end site"))Metagene plot
ggplot(features, aes(x = pos, y = rpm, color = sample_id)) +
geom_line() +
facet_wrap(~feature) +
scale_color_discrete(name = "Sample") +
labs(x = "Relative position (bp)",
y = "Counts per million (mean)",
title = "5' bias of UMI protocol") +
theme(legend.position = "bottom")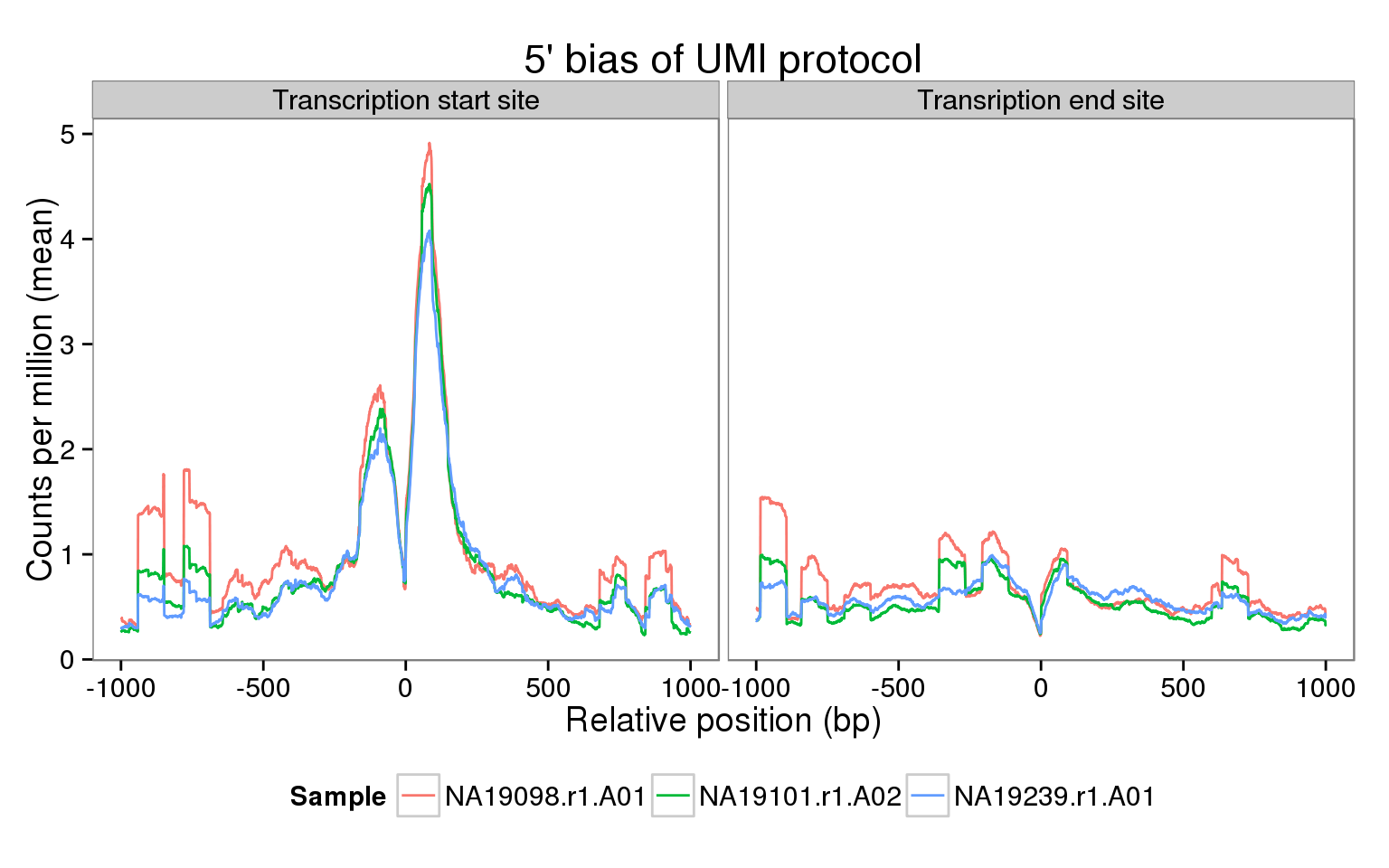
Interpretation
These results are similar to those obtained using the molecules. Furthermore, they have many more reads and thus do not suffer as much from sparsity.
reads_filter_sub <- reads_filter[, names(quality_cells)]
colSums(reads_filter_sub) / 10^3NA19098.r1.A01 NA19101.r1.A02 NA19239.r1.A01
1772.526 2754.120 2164.597 mean_expr <- rowMeans(reads_filter_sub)
summary(mean_expr) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 1.00 46.33 182.30 143.70 22910.00 The median across the genes for the mean number of molecules across these three samples is 46.3333333 molecules.
Session information
sessionInfo()R version 3.2.0 (2015-04-16)
Platform: x86_64-unknown-linux-gnu (64-bit)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 grid stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] ggplot2_1.0.1 tidyr_0.2.0 plyr_1.8.3
[4] Rsamtools_1.20.4 Biostrings_2.36.1 XVector_0.8.0
[7] GenomicRanges_1.20.5 GenomeInfoDb_1.4.0 IRanges_2.2.4
[10] S4Vectors_0.6.0 BiocGenerics_0.14.0 genomation_1.0.0
[13] knitr_1.10.5
loaded via a namespace (and not attached):
[1] Rcpp_0.12.0 formatR_1.2
[3] futile.logger_1.4.1 bitops_1.0-6
[5] futile.options_1.0.0 tools_3.2.0
[7] zlibbioc_1.14.0 digest_0.6.8
[9] evaluate_0.7 gtable_0.1.2
[11] gridBase_0.4-7 DBI_0.3.1
[13] yaml_2.1.13 proto_0.3-10
[15] dplyr_0.4.2 rtracklayer_1.28.4
[17] httr_0.6.1 stringr_1.0.0
[19] data.table_1.9.4 impute_1.42.0
[21] R6_2.1.1 XML_3.98-1.2
[23] BiocParallel_1.2.2 rmarkdown_0.6.1
[25] reshape2_1.4.1 lambda.r_1.1.7
[27] magrittr_1.5 codetools_0.2-11
[29] scales_0.2.4 htmltools_0.2.6
[31] GenomicAlignments_1.4.1 MASS_7.3-40
[33] assertthat_0.1 colorspace_1.2-6
[35] labeling_0.3 stringi_0.4-1
[37] lazyeval_0.1.10 RCurl_1.95-4.6
[39] munsell_0.4.2 chron_2.3-45