Per gene individual differences in CV and mean
Joyce Hsiao
2015-10-23
Last updated: 2015-12-11
Code version: 884b335605607bd2cf5c9a1ada1e345466388866
Objective
Previously, we compared normalized coefficient of variations across individuals. Here, we will also compare the mean gene expression across individuals, using single cell sequencing data and bulk RNA-seq data.
It would be interesting to learn the possible overlap or non-overlap between the genes that we observed significant individual differences in coefficient of variations versus those different in mean gene expression levels across cells.
From here on we will work with the filtered data without NA19098.r2.
Set up
library("data.table")
library("dplyr")
library("limma")
library("edgeR")
library("ggplot2")
library("grid")
theme_set(theme_bw(base_size = 12))
source("functions.R")Prepare data
Input annotation of only QC-filtered single cells. Remove NA19098.r2
anno_qc_filter <- read.table("../data/annotation-filter.txt", header = TRUE,
stringsAsFactors = FALSE)Import endogeneous gene molecule counts that are QC-filtered, CPM-normalized, ERCC-normalized, and also processed to remove unwanted variation from batch effet. ERCC genes are removed from this file.
molecules_ENSG <- read.table("../data/molecules-final.txt", header = TRUE, stringsAsFactors = FALSE)Input moleclule counts before log2 CPM transformation. This file is used to compute percent zero-count cells per sample.
molecules_sparse <- read.table("../data/molecules-filter.txt", header = TRUE, stringsAsFactors = FALSE)
molecules_sparse <- molecules_sparse[grep("ENSG", rownames(molecules_sparse)), ]
stopifnot( all.equal(rownames(molecules_ENSG), rownames(molecules_sparse)) )Normalize coefficient of variation
Compute coefficient of variation.
# Compute CV and mean of normalized molecule counts (take 2^(log2-normalized count))
molecules_cv_batch_ENSG <-
lapply(1:length(unique(anno_qc_filter$batch)), function(per_batch) {
molecules_per_batch <- 2^molecules_ENSG[ , unique(anno_qc_filter$batch) == unique(anno_qc_filter$batch)[per_batch] ]
mean_per_gene <- apply(molecules_per_batch, 1, mean, na.rm = TRUE)
sd_per_gene <- apply(molecules_per_batch, 1, sd, na.rm = TRUE)
cv_per_gene <- data.frame(mean = mean_per_gene,
sd = sd_per_gene,
cv = sd_per_gene/mean_per_gene)
rownames(cv_per_gene) <- rownames(molecules_ENSG)
# cv_per_gene <- cv_per_gene[rowSums(is.na(cv_per_gene)) == 0, ]
cv_per_gene$batch <- unique(anno_qc_filter$batch)[per_batch]
# Add sparsity percent
molecules_sparse_per_batch <- molecules_sparse[ , unique(anno_qc_filter$batch) == unique(anno_qc_filter$batch)[per_batch]]
cv_per_gene$sparse <- rowMeans(as.matrix(molecules_sparse_per_batch) == 0)
return(cv_per_gene)
})
names(molecules_cv_batch_ENSG) <- unique(anno_qc_filter$batch)Merge summary data.frames.
df_ENSG <- do.call(rbind, molecules_cv_batch_ENSG)Compute rolling medians across all samples.
library(zoo)
# Compute a data-wide coefficient of variation on CPM normalized counts.
data_cv_ENSG <- apply(2^molecules_ENSG, 1, sd)/apply(2^molecules_ENSG, 1, mean)
# Order of genes by mean expression levels
order_gene <- order(apply(2^molecules_ENSG, 1, mean))
# Rolling medians of log10 squared CV by mean expression levels
roll_medians <- rollapply(log10(data_cv_ENSG^2)[order_gene], width = 50, by = 25,
FUN = median, fill = list("extend", "extend", "NA") )
ii_na <- which( is.na(roll_medians) )
roll_medians[ii_na] <- median( log10(data_cv_ENSG^2)[order_gene][ii_na] )
names(roll_medians) <- rownames(molecules_ENSG)[order_gene]
# re-order rolling medians
reorder_gene <- match(rownames(molecules_ENSG), names(roll_medians) )
roll_medians <- roll_medians[ reorder_gene ]
stopifnot( all.equal(names(roll_medians), rownames(molecules_ENSG) ) )Compute adjusted coefficient of variation.
# adjusted coefficient of variation on log10 scale
log10cv2_adj_ENSG <-
lapply(1:length(molecules_cv_batch_ENSG), function(per_batch) {
foo <- log10(molecules_cv_batch_ENSG[[per_batch]]$cv^2) - roll_medians
return(foo)
})
df_ENSG$log10cv2_adj_ENSG <- do.call(c, log10cv2_adj_ENSG)Compare coefficients of variations
library(limma)
df_limma <- matrix(df_ENSG$log10cv2_adj_ENSG,
nrow = nrow(molecules_ENSG), ncol = 8, byrow = FALSE)
design <- data.frame(individual = factor(rep(unique(anno_qc_filter$individual), each = 3) ),
rep = factor(rep(c(1:3), times = 3)) )
design <- design[ with(design, !(individual == "NA19098" & rep == "2")), ]
colnames(df_limma) <- with(design, paste0(individual, rep))
fit_limma <- lmFit(df_limma, design = model.matrix( ~ individual, data = design))
fit_limma <- eBayes(fit_limma)False discovery control adjustment.
F.p.adj <- p.adjust(fit_limma$F.p.value, method = "fdr")Cutoffs
df_cuts <- data.frame(cuts = c(.001, .01, .05, .1, .15, .2))
df_cuts$sig_count <- sapply(1:6, function(per_cut) {
sum(F.p.adj < df_cuts$cuts[per_cut] )
})
df_cuts cuts sig_count
1 0.001 91
2 0.010 192
3 0.050 385
4 0.100 560
5 0.150 780
6 0.200 1004False discovery control adjutment.
F.p.adj <- p.adjust(fit_limma$F.p.value, method = "fdr")Squared CV vs. mean expression level
We plotted out per gene average CVs across samples versus average expression level for the endogeneous genes, and identified the genes classifed as significantly different between individuals in coefficients of variation.
df_compare <-
data.frame(mean = rowMeans( as.matrix(
do.call(cbind, lapply(molecules_cv_batch_ENSG, "[[", 1) ) ) ),
cv2 = rowMeans( as.matrix(
do.call(cbind, lapply(molecules_cv_batch_ENSG, "[[", 3) ) )^2 ),
adj_cv2 = rowMeans( 10^as.matrix(
do.call(cbind, log10cv2_adj_ENSG) ) ) )
library(broman)
crayon <- brocolors("crayons")
par(mar=c(5,5,3,1))
with(df_compare, plot(x = log10(mean), y = cv2, pch = 1, cex = 1, col = "grey50",
lwd = .5,
ylab = "average squared coeffcient of variations \n across samples",
xlab = "log10 average of mean CPM across samples") )
with(df_compare[F.p.adj < .2, ], points(x = log10(mean), y = cv2, pch = 16, cex = .6,
col = crayon["Tumbleweed"]) )
with(df_compare[F.p.adj < .1, ], points(x = log10(mean), y = cv2, pch = 16, cex = .6,
col = crayon["Orange"]) )
with(df_compare[F.p.adj < .01, ], points(x = log10(mean), y = cv2, pch = 16, cex = .6,
col = crayon["Scarlet"]) )
title(main = "Avg. Squred CV vs. log10(Avg. mean count)")
legend("topright", pch = c(1, 16, 16, 16),
legend = c("All genes", "Adj p-value < .2",
"Adj p-value < .1", "Adj p-value < .01"),
col = c("grey50", crayon[c("Tumbleweed", "Orange", "Scarlet")]),
bty = "n")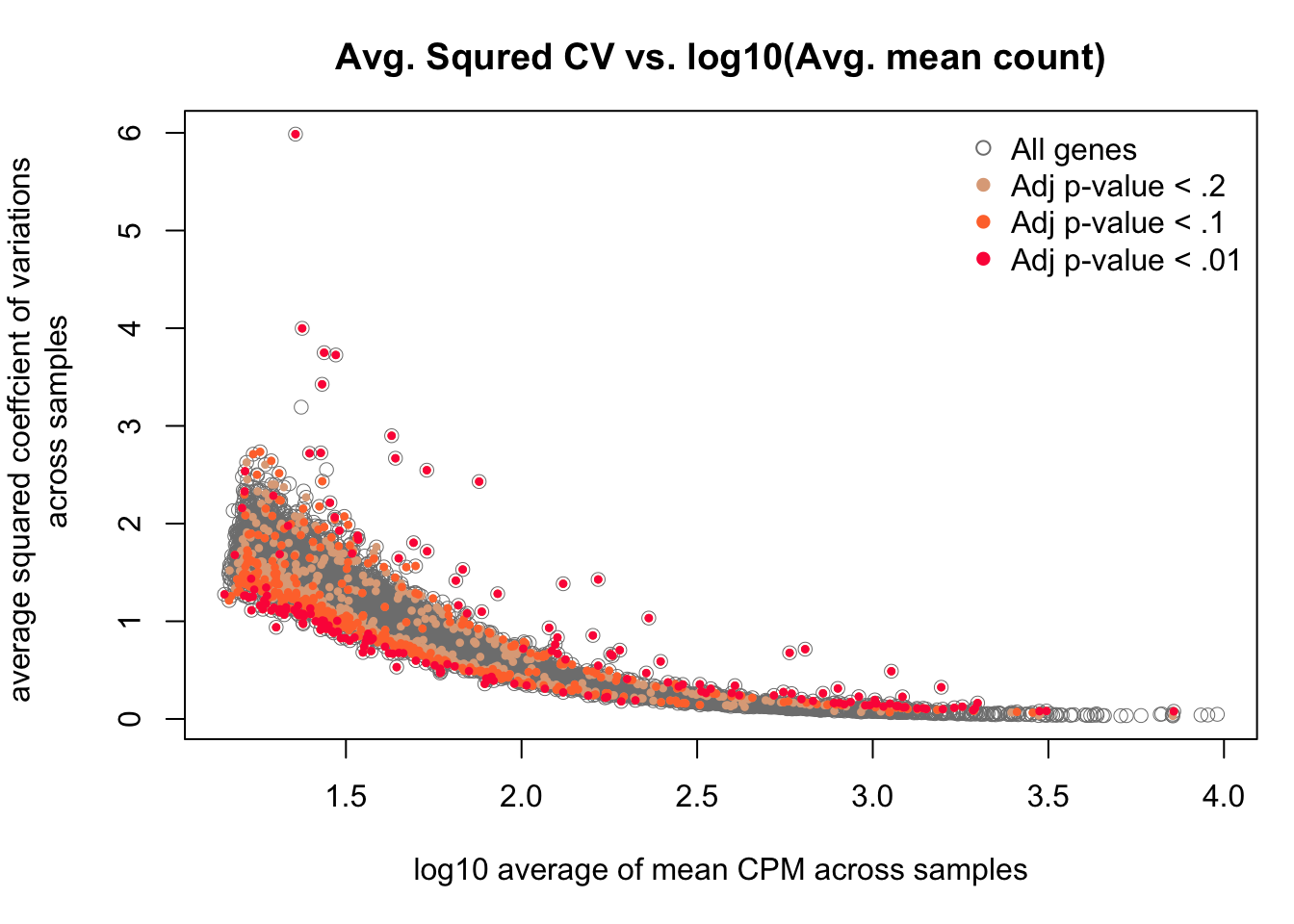
par(mar=c(5,5,3,1))
with(df_compare, plot(x = log10(mean), y = adj_cv2, pch = 1, cex = 1, col = "grey50",
lwd = .5,
ylab = "average squared coeffcient of variations \n across samples",
xlab = "log10 average of mean CPM across samples") )
with(df_compare[F.p.adj < .2, ], points(x = log10(mean), y = adj_cv2, pch = 16, cex = .6,
col = crayon["Tumbleweed"]) )
with(df_compare[F.p.adj < .1, ], points(x = log10(mean), y = adj_cv2, pch = 16, cex = .6,
col = crayon["Orange"]) )
with(df_compare[F.p.adj < .01, ], points(x = log10(mean), y = adj_cv2, pch = 16, cex = .6,
col = crayon["Scarlet"]) )
title(main = "Avg. Adjusted Squred CV vs. log10(Avg. mean count)")
legend("topright", pch = c(1, 16, 16, 16),
legend = c("All genes", "Adj p-value < .2",
"Adj p-value < .1", "Adj p-value < .01"),
col = c("grey50", crayon[c("Tumbleweed", "Orange", "Scarlet")]),
bty = "n")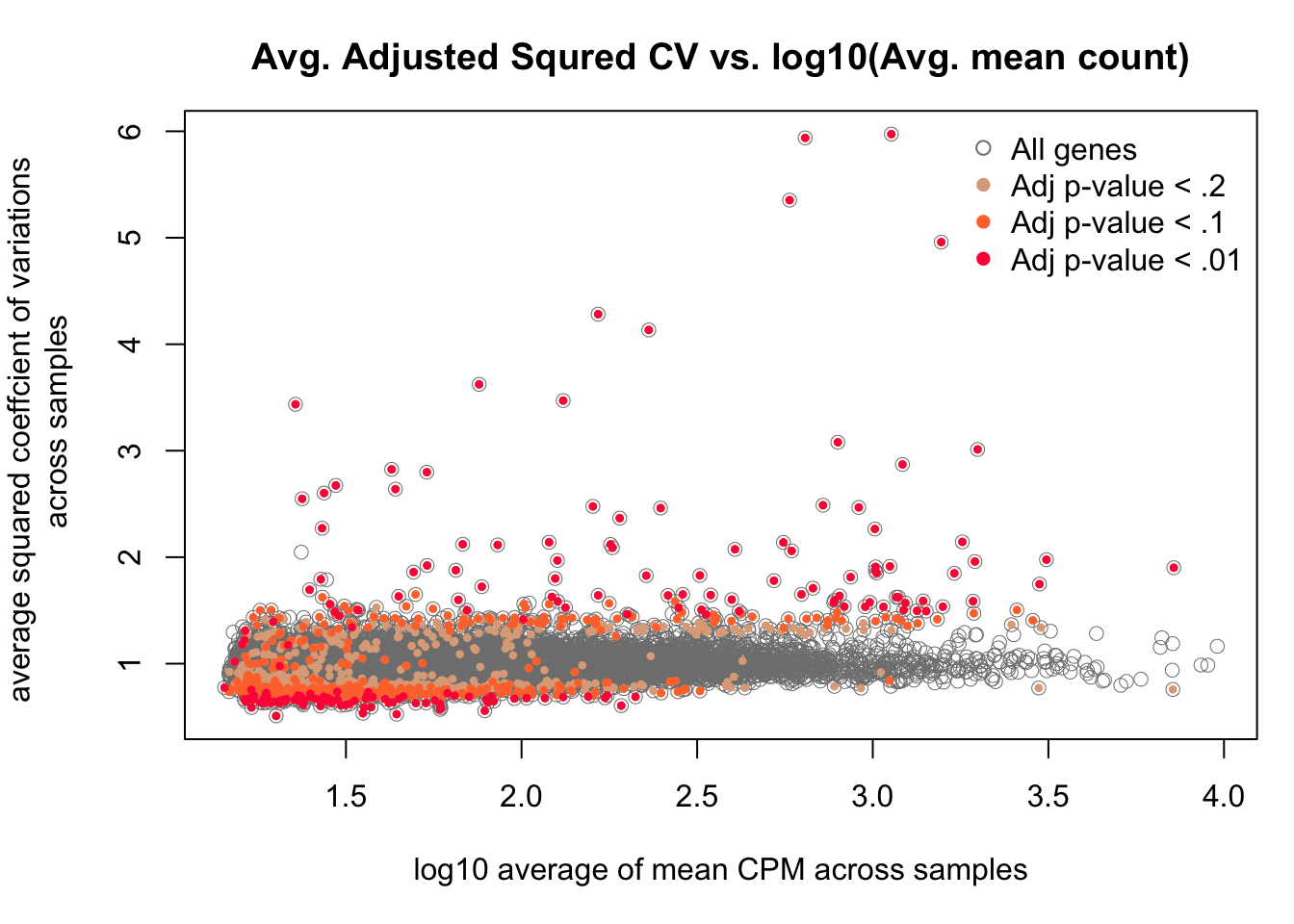
Investigate…
Our naive analysis seems to pick up genes with different density between individuals.
library(gridExtra)
order_low_cv <- order(df_compare[F.p.adj < .01, ]$cv)
low_plots <- lapply(1:6, function(i) {
ind <- order_low_cv[i]
ggplot(data.frame(values = unlist(molecules_ENSG[ind, ]),
individual = factor(anno_qc_filter$individual),
replicate = factor(anno_qc_filter$replicate),
batch = factor(anno_qc_filter$batch), check.rows = F),
aes(x = values)) +
geom_density(aes(group = batch, col = individual)) +
ggtitle(paste(rownames(molecules_ENSG)[ind],
";CV = ", round(df_compare[F.p.adj < .01, ]$cv[ind], 2) ) )
})
grid.arrange(grobs = low_plots, ncol = 2)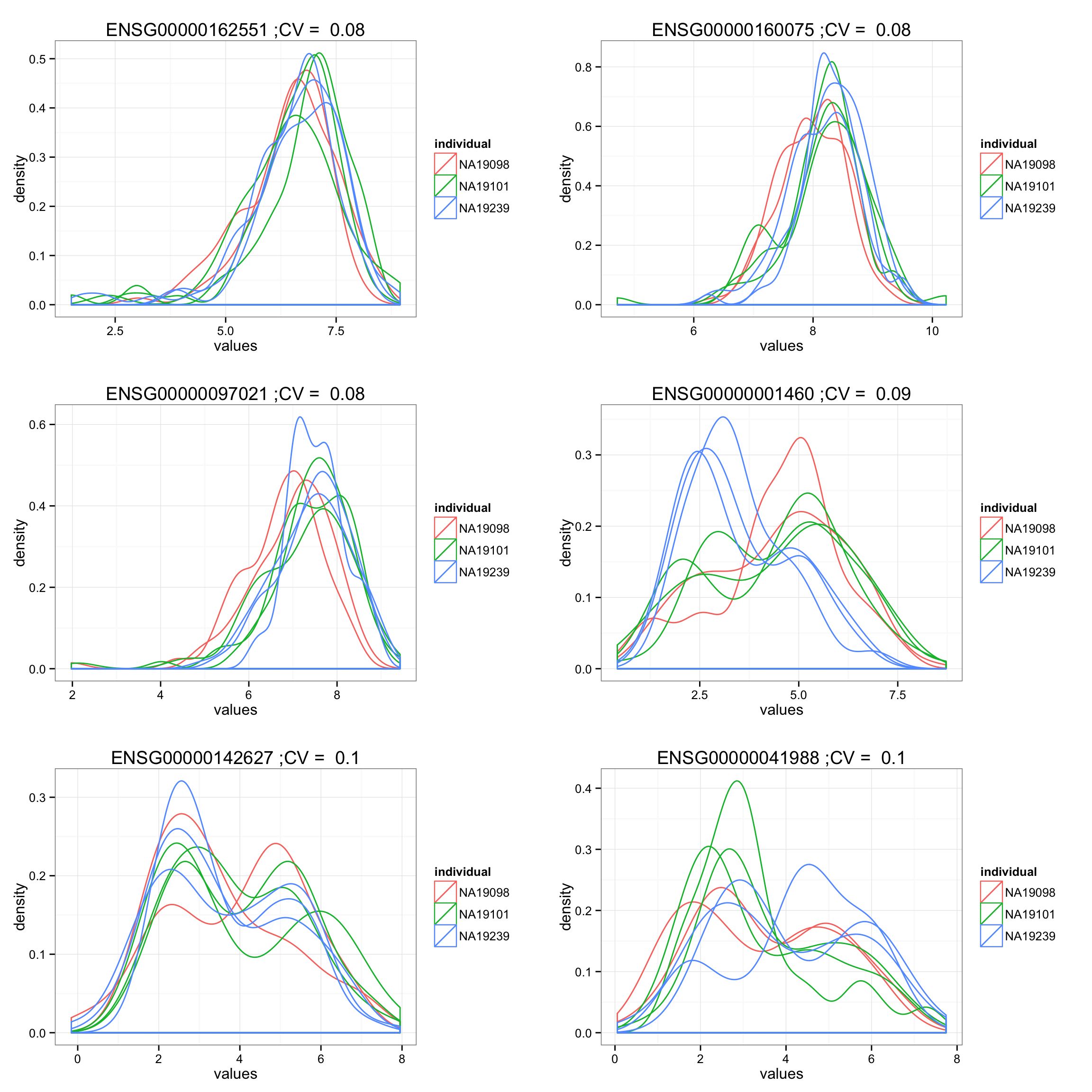
order_high_cv <- order(df_compare[F.p.adj < .01, ]$cv, decreasing = TRUE)
high_plots <- lapply(1:6, function(i) {
ind <- order_high_cv[i]
ggplot(data.frame(values = unlist(molecules_ENSG[ind, ]),
individual = factor(anno_qc_filter$individual),
replicate = factor(anno_qc_filter$replicate),
batch = factor(anno_qc_filter$batch), check.rows = F),
aes(x = values)) +
geom_density(aes(group = batch, col = individual)) +
ggtitle(paste(rownames(molecules_ENSG)[ind],
";CV = ", round(df_compare[F.p.adj < .01, ]$cv[ind], 2) ) )
})
grid.arrange(grobs = high_plots, ncol = 2)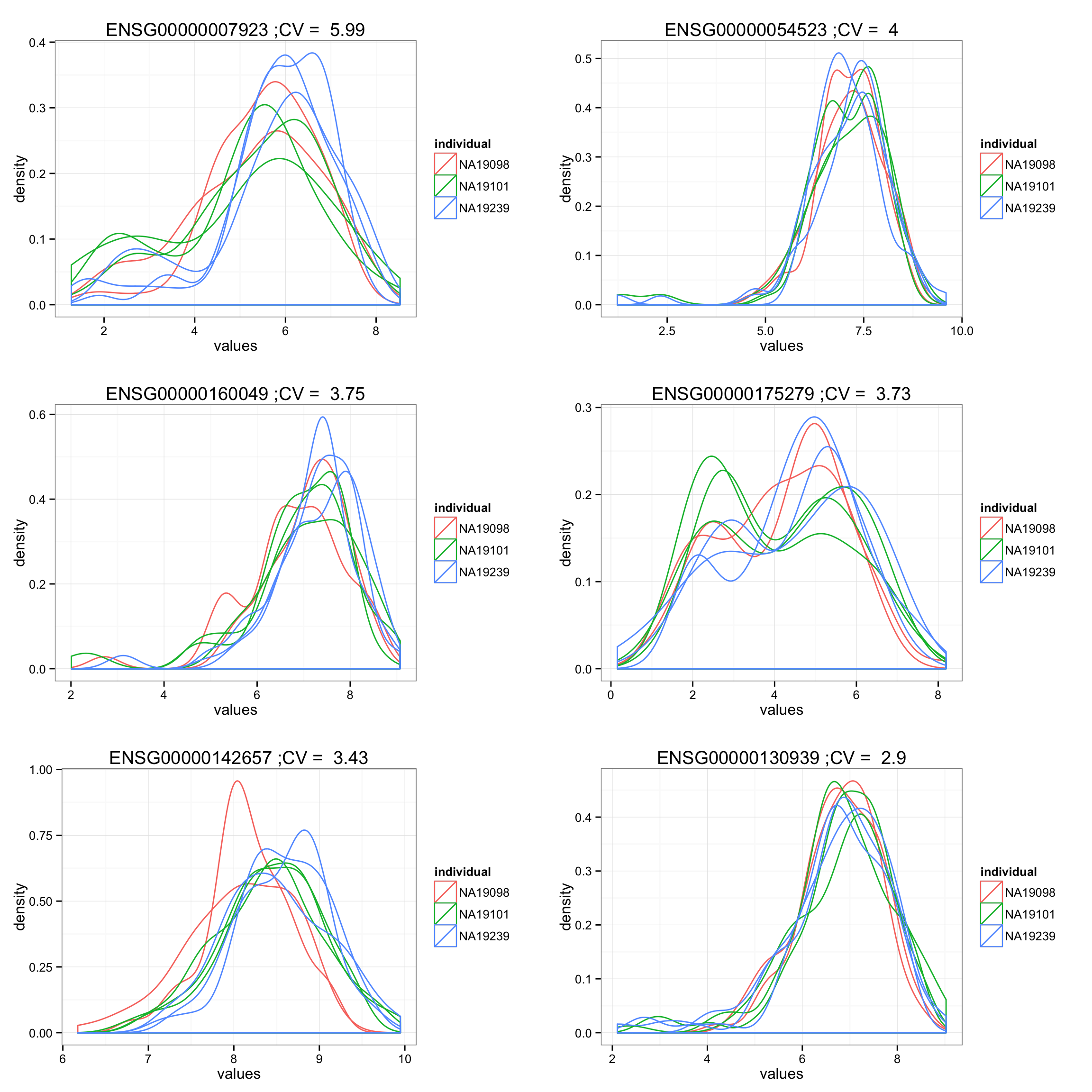
Annotation
ENSEMBLE database
sig_cv <- order(F.p.adj)[F.p.adj < .05]
library("biomaRt")
ensembl <- useMart(host = "grch37.ensembl.org",
biomart = "ENSEMBL_MART_ENSEMBL",
dataset = "hsapiens_gene_ensembl")
differential_CV_genes_info <- getBM(attributes = c("ensembl_gene_id", "chromosome_name",
"external_gene_name", "transcript_count",
"description"),
filters = "ensembl_gene_id",
values = rownames(molecules_ENSG[sig_cv, ]),
mart = ensembl)
differential_CV_genes_info ensembl_gene_id chromosome_name external_gene_name transcript_count
1 ENSG00000006432 14 MAP3K9 6
2 ENSG00000008018 6 PSMB1 2
3 ENSG00000008311 7 AASS 7
4 ENSG00000008988 8 RPS20 11
5 ENSG00000010072 1 SPRTN 6
6 ENSG00000011021 1 CLCN6 9
7 ENSG00000011114 14 BTBD7 9
8 ENSG00000013561 5 RNF14 18
9 ENSG00000029725 17 RABEP1 10
10 ENSG00000030419 2 IKZF2 17
11 ENSG00000033170 14 FUT8 17
12 ENSG00000050393 6 MCUR1 4
13 ENSG00000051825 12 MPHOSPH9 26
14 ENSG00000060339 10 CCAR1 18
15 ENSG00000062485 12 CS 33
16 ENSG00000063177 19 RPL18 16
17 ENSG00000064995 6 TAF11 2
18 ENSG00000065548 2 ZC3H15 10
19 ENSG00000066136 1 NFYC 24
20 ENSG00000068400 X GRIPAP1 9
21 ENSG00000069943 15 PIGB 12
22 ENSG00000070756 8 PABPC1 23
23 ENSG00000071054 2 MAP4K4 18
24 ENSG00000073111 3 MCM2 9
25 ENSG00000075234 22 TTC38 7
26 ENSG00000076382 17 SPAG5 16
27 ENSG00000076706 11 MCAM 18
28 ENSG00000077713 X SLC25A43 5
29 ENSG00000078967 7 UBE2D4 12
30 ENSG00000079215 5 SLC1A3 14
31 ENSG00000079277 1 MKNK1 30
32 ENSG00000079739 1 PGM1 5
33 ENSG00000080371 12 RAB21 4
34 ENSG00000081692 1 JMJD4 5
35 ENSG00000085733 11 CTTN 16
36 ENSG00000087008 4 ACOX3 8
37 ENSG00000087157 17 PGS1 16
38 ENSG00000088035 1 ALG6 7
39 ENSG00000090060 14 PAPOLA 25
40 ENSG00000090971 19 NAT14 5
41 ENSG00000091039 12 OSBPL8 17
42 ENSG00000092330 14 TINF2 16
43 ENSG00000093144 6 ECHDC1 24
44 ENSG00000097021 1 ACOT7 11
45 ENSG00000099139 9 PCSK5 5
46 ENSG00000100106 22 TRIOBP 11
47 ENSG00000100129 22 EIF3L 13
48 ENSG00000100139 22 MICALL1 6
49 ENSG00000100296 22 THOC5 17
50 ENSG00000100629 14 CEP128 17
51 ENSG00000100644 14 HIF1A 12
52 ENSG00000100784 14 RPS6KA5 7
53 ENSG00000100908 14 EMC9 7
54 ENSG00000100916 14 BRMS1L 7
55 ENSG00000101189 20 MRGBP 1
56 ENSG00000101194 20 SLC17A9 7
57 ENSG00000101198 20 NKAIN4 9
58 ENSG00000101333 20 PLCB4 14
59 ENSG00000101493 18 ZNF516 6
60 ENSG00000101596 18 SMCHD1 14
61 ENSG00000101974 X ATP11C 11
62 ENSG00000101997 X CCDC22 3
63 ENSG00000102054 X RBBP7 12
64 ENSG00000102078 X SLC25A14 14
65 ENSG00000102158 X MAGT1 3
66 ENSG00000102189 12 EEA1 5
67 ENSG00000102226 X USP11 12
68 ENSG00000102531 13 FNDC3A 6
69 ENSG00000102901 16 CENPT 36
70 ENSG00000103316 16 CRYM 12
71 ENSG00000103356 16 EARS2 17
72 ENSG00000103449 16 SALL1 6
73 ENSG00000103544 16 C16orf62 25
74 ENSG00000103978 15 TMEM87A 13
75 ENSG00000104497 8 SNX16 11
76 ENSG00000104611 8 SH2D4A 6
77 ENSG00000104885 19 DOT1L 11
78 ENSG00000105325 19 FZR1 8
79 ENSG00000105520 19 DKFZP761J1410 8
80 ENSG00000106236 7 NPTX2 2
81 ENSG00000106785 9 TRIM14 7
82 ENSG00000107021 9 TBC1D13 5
83 ENSG00000108010 10 GLRX3 5
84 ENSG00000108176 10 DNAJC12 5
85 ENSG00000108384 17 RAD51C 17
86 ENSG00000108774 17 RAB5C 9
87 ENSG00000109255 4 NMU 6
88 ENSG00000109576 4 AADAT 8
89 ENSG00000109670 4 FBXW7 13
90 ENSG00000109689 4 STIM2 13
91 ENSG00000110046 11 ATG2A 6
92 ENSG00000110063 11 DCPS 3
93 ENSG00000110074 11 FOXRED1 19
94 ENSG00000110422 11 HIPK3 6
95 ENSG00000110514 11 MADD 22
96 ENSG00000111087 12 GLI1 8
97 ENSG00000111215 12 PRR4 22
98 ENSG00000111271 12 ACAD10 25
99 ENSG00000111450 12 STX2 4
100 ENSG00000111670 12 GNPTAB 10
101 ENSG00000111911 6 HINT3 1
102 ENSG00000112081 6 SRSF3 3
103 ENSG00000112305 6 SMAP1 6
104 ENSG00000112699 6 GMDS 10
105 ENSG00000112715 6 VEGFA 25
106 ENSG00000113758 5 DBN1 12
107 ENSG00000114388 3 NPRL2 17
108 ENSG00000114784 3 EIF1B 4
109 ENSG00000115539 2 PDCL3 3
110 ENSG00000115738 2 ID2 4
111 ENSG00000115841 2 RMDN2 12
112 ENSG00000116285 1 ERRFI1 5
113 ENSG00000116641 1 DOCK7 8
114 ENSG00000116704 1 SLC35D1 2
115 ENSG00000116991 1 SIPA1L2 6
116 ENSG00000117020 1 AKT3 10
117 ENSG00000117280 1 RAB7L1 9
118 ENSG00000118200 1 CAMSAP2 6
119 ENSG00000118564 4 FBXL5 14
120 ENSG00000119599 14 DCAF4 15
121 ENSG00000119686 14 FLVCR2 13
122 ENSG00000119866 2 BCL11A 12
123 ENSG00000120053 10 GOT1 4
124 ENSG00000120526 8 NUDCD1 4
125 ENSG00000120756 3 PLS1 12
126 ENSG00000121057 17 AKAP1 18
127 ENSG00000121644 1 DESI2 4
128 ENSG00000121680 11 PEX16 11
129 ENSG00000123066 12 MED13L 10
130 ENSG00000123179 13 EBPL 8
131 ENSG00000123219 5 CENPK 17
132 ENSG00000123384 12 LRP1 12
133 ENSG00000123643 5 SLC36A1 10
134 ENSG00000123933 4 MXD4 5
135 ENSG00000124145 20 SDC4 2
136 ENSG00000124493 6 GRM4 14
137 ENSG00000124508 6 BTN2A2 16
138 ENSG00000125398 17 SOX9 1
139 ENSG00000125449 17 ARMC7 5
140 ENSG00000125505 19 MBOAT7 14
141 ENSG00000125686 17 MED1 5
142 ENSG00000125817 20 CENPB 1
143 ENSG00000125845 20 BMP2 1
144 ENSG00000126107 1 HECTD3 7
145 ENSG00000126790 14 L3HYPDH 9
146 ENSG00000126858 17 RHOT1 17
147 ENSG00000127125 1 PPCS 9
148 ENSG00000129055 3 ANAPC13 6
149 ENSG00000129317 12 PUS7L 8
150 ENSG00000129353 19 SLC44A2 15
151 ENSG00000130340 6 SNX9 1
152 ENSG00000130559 9 CAMSAP1 10
153 ENSG00000130764 1 LRRC47 3
154 ENSG00000131149 16 GSE1 12
155 ENSG00000131153 16 GINS2 3
156 ENSG00000131876 15 SNRPA1 15
157 ENSG00000132405 4 TBC1D14 9
158 ENSG00000132475 17 H3F3B 13
159 ENSG00000132561 8 MATN2 18
160 ENSG00000132846 5 ZBED3 3
161 ENSG00000133169 X BEX1 1
162 ENSG00000133226 1 SRRM1 24
163 ENSG00000133243 19 BTBD2 10
164 ENSG00000133313 18 CNDP2 29
165 ENSG00000134049 18 IER3IP1 1
166 ENSG00000134198 1 TSPAN2 5
167 ENSG00000134247 1 PTGFRN 3
168 ENSG00000134318 2 ROCK2 8
169 ENSG00000134363 5 FST 5
170 ENSG00000135272 7 MDFIC 7
171 ENSG00000135473 12 PAN2 20
172 ENSG00000135486 12 HNRNPA1 14
173 ENSG00000136261 7 BZW2 13
174 ENSG00000136536 2 MARCH7 9
175 ENSG00000136560 2 TANK 24
176 ENSG00000136717 2 BIN1 14
177 ENSG00000136868 9 SLC31A1 3
178 ENSG00000137040 9 RANBP6 2
179 ENSG00000137331 6 IER3 2
180 ENSG00000137364 6 TPMT 1
181 ENSG00000137947 1 GTF2B 7
182 ENSG00000138018 2 EPT1 4
183 ENSG00000138107 10 ACTR1A 8
184 ENSG00000138622 15 HCN4 1
185 ENSG00000138944 22 KIAA1644 1
186 ENSG00000139679 13 LPAR6 8
187 ENSG00000140395 15 WDR61 16
188 ENSG00000140830 16 TXNL4B 6
189 ENSG00000140832 16 MARVELD3 6
190 ENSG00000140905 16 GCSH 6
191 ENSG00000140941 16 MAP1LC3B 10
192 ENSG00000140988 16 RPS2 17
193 ENSG00000141002 16 TCF25 24
194 ENSG00000141738 17 GRB7 15
195 ENSG00000142065 19 ZFP14 2
196 ENSG00000142168 21 SOD1 4
197 ENSG00000142530 19 FAM71E1 6
198 ENSG00000142541 19 RPL13A 12
199 ENSG00000142634 1 EFHD2 2
200 ENSG00000143320 1 CRABP2 3
201 ENSG00000143353 1 LYPLAL1 12
202 ENSG00000143776 1 CDC42BPA 15
203 ENSG00000144034 2 TPRKB 10
204 ENSG00000144233 2 AMMECR1L 2
205 ENSG00000144401 2 METTL21A 13
206 ENSG00000144711 3 IQSEC1 5
207 ENSG00000144827 3 ABHD10 6
208 ENSG00000145016 3 KIAA0226 11
209 ENSG00000145293 4 ENOPH1 3
210 ENSG00000145632 5 PLK2 14
211 ENSG00000145730 5 PAM 29
212 ENSG00000145912 5 NHP2 6
213 ENSG00000146350 6 TBC1D32 9
214 ENSG00000146433 6 TMEM181 1
215 ENSG00000146856 7 AGBL3 9
216 ENSG00000147027 X TMEM47 1
217 ENSG00000147394 X ZNF185 12
218 ENSG00000147416 8 ATP6V1B2 6
219 ENSG00000147854 9 UHRF2 13
220 ENSG00000147905 9 ZCCHC7 9
221 ENSG00000148297 9 MED22 10
222 ENSG00000148300 9 REXO4 6
223 ENSG00000148337 9 CIZ1 27
224 ENSG00000148399 9 DPH7 14
225 ENSG00000148908 10 RGS10 5
226 ENSG00000149131 11 SERPING1 12
227 ENSG00000149743 11 TRPT1 16
228 ENSG00000150471 4 LPHN3 19
229 ENSG00000151413 14 NUBPL 14
230 ENSG00000152904 1 GGPS1 9
231 ENSG00000152939 5 MARVELD2 6
232 ENSG00000153989 6 NUS1 1
233 ENSG00000154545 X MAGED4 11
234 ENSG00000154556 4 SORBS2 65
235 ENSG00000154620 Y TMSB4Y 1
236 ENSG00000155393 16 HEATR3 6
237 ENSG00000155975 8 VPS37A 12
238 ENSG00000156931 3 VPS8 27
239 ENSG00000157014 3 TATDN2 4
240 ENSG00000157823 15 AP3S2 15
241 ENSG00000157837 12 SPPL3 9
242 ENSG00000157851 2 DPYSL5 6
243 ENSG00000158006 1 PAFAH2 7
244 ENSG00000158079 9 PTPDC1 3
245 ENSG00000158864 1 NDUFS2 16
246 ENSG00000159202 17 UBE2Z 8
247 ENSG00000159459 15 UBR1 12
248 ENSG00000160075 1 SSU72 4
249 ENSG00000160131 X VMA21 3
250 ENSG00000160214 21 RRP1 7
251 ENSG00000160551 17 TAOK1 6
252 ENSG00000160584 11 SIK3 17
253 ENSG00000161551 19 ZNF577 17
254 ENSG00000161958 17 FGF11 6
255 ENSG00000161996 16 WDR90 25
256 ENSG00000162231 11 NXF1 18
257 ENSG00000162576 1 MXRA8 10
258 ENSG00000162616 1 DNAJB4 6
259 ENSG00000162836 1 ACP6 6
260 ENSG00000162998 2 FRZB 1
261 ENSG00000163125 1 RPRD2 5
262 ENSG00000163362 1 C1orf106 7
263 ENSG00000163428 3 LRRC58 1
264 ENSG00000163482 2 STK36 17
265 ENSG00000163584 3 RPL22L1 6
266 ENSG00000163704 3 PRRT3 3
267 ENSG00000163930 3 BAP1 10
268 ENSG00000163947 3 ARHGEF3 15
269 ENSG00000164056 4 SPRY1 6
270 ENSG00000164081 3 TEX264 14
271 ENSG00000164117 4 FBXO8 4
272 ENSG00000164163 4 ABCE1 10
273 ENSG00000164663 6 USP49 8
274 ENSG00000164764 8 SBSPON 2
275 ENSG00000164933 8 SLC25A32 6
276 ENSG00000165169 X DYNLT3 3
277 ENSG00000165275 9 TRMT10B 9
278 ENSG00000165630 10 PRPF18 3
279 ENSG00000165916 11 PSMC3 12
280 ENSG00000166426 15 CRABP1 3
281 ENSG00000166704 19 ZNF606 10
282 ENSG00000166888 12 STAT6 24
283 ENSG00000167118 9 URM1 6
284 ENSG00000167302 17 ENTHD2 13
285 ENSG00000167306 18 MYO5B 8
286 ENSG00000167380 19 ZNF226 19
287 ENSG00000167654 19 ATCAY 8
288 ENSG00000168040 11 FADD 1
289 ENSG00000168872 16 DDX19A 16
290 ENSG00000169021 19 UQCRFS1 1
291 ENSG00000169169 19 CPT1C 25
292 ENSG00000169814 3 BTD 12
293 ENSG00000169905 1 TOR1AIP2 6
294 ENSG00000169908 3 TM4SF1 4
295 ENSG00000169951 16 ZNF764 3
296 ENSG00000170266 3 GLB1 20
297 ENSG00000170448 4 NFXL1 8
298 ENSG00000170871 4 KIAA0232 5
299 ENSG00000171161 1 ZNF672 7
300 ENSG00000171310 12 CHST11 6
301 ENSG00000171700 20 RGS19 4
302 ENSG00000171861 17 RNMTL1 4
303 ENSG00000172167 8 MTBP 8
304 ENSG00000172717 14 FAM71D 10
305 ENSG00000173465 11 SSSCA1 7
306 ENSG00000173486 11 FKBP2 6
307 ENSG00000173744 2 AGFG1 8
308 ENSG00000173926 5 MARCH3 5
309 ENSG00000174456 12 C12orf76 8
310 ENSG00000174695 5 TMEM167A 6
311 ENSG00000175130 1 MARCKSL1 1
312 ENSG00000175215 12 CTDSP2 8
313 ENSG00000175279 1 APITD1 7
314 ENSG00000177105 11 RHOG 4
315 ENSG00000177119 12 ANO6 8
316 ENSG00000177600 11 RPLP2 8
317 ENSG00000177879 5 AP3S1 8
318 ENSG00000179943 19 FIZ1 5
319 ENSG00000179958 16 DCTPP1 5
320 ENSG00000180822 6 PSMG4 10
321 ENSG00000181827 15 RFX7 6
322 ENSG00000181830 11 SLC35C1 5
323 ENSG00000181915 10 ADO 1
324 ENSG00000182919 11 C11orf54 14
325 ENSG00000182986 19 ZNF320 14
326 ENSG00000183530 22 PRR14L 9
327 ENSG00000183864 22 TOB2 2
328 ENSG00000184007 1 PTP4A2 20
329 ENSG00000184348 6 HIST1H2AK 1
330 ENSG00000185215 14 TNFAIP2 13
331 ENSG00000185420 1 SMYD3 19
332 ENSG00000185684 12 EP400NL 14
333 ENSG00000185721 22 DRG1 6
334 ENSG00000185722 17 ANKFY1 14
335 ENSG00000186635 11 ARAP1 21
336 ENSG00000186716 22 BCR 16
337 ENSG00000187140 1 FOXD3 1
338 ENSG00000187193 16 MT1X 4
339 ENSG00000187514 2 PTMA 12
340 ENSG00000188690 10 UROS 10
341 ENSG00000189334 1 S100A14 6
342 ENSG00000196374 6 HIST1H2BM 1
343 ENSG00000196648 15 GOLGA6L20 4
344 ENSG00000197102 14 DYNC1H1 11
345 ENSG00000197183 20 C20orf112 9
346 ENSG00000197459 6 HIST1H2BH 1
347 ENSG00000197879 17 MYO1C 21
348 ENSG00000198176 13 TFDP1 9
349 ENSG00000198315 6 ZKSCAN8 4
350 ENSG00000198393 12 ZNF26 6
351 ENSG00000198464 19 ZNF480 6
352 ENSG00000198589 4 LRBA 14
353 ENSG00000198612 2 COPS8 8
354 ENSG00000198887 9 SMC5 3
355 ENSG00000198917 9 C9orf114 5
356 ENSG00000198960 X ARMCX6 8
357 ENSG00000203930 X LINC00632 5
358 ENSG00000204071 X TCEAL6 2
359 ENSG00000204356 6 NELFE 13
360 ENSG00000204442 13 FAM155A 1
361 ENSG00000204536 6 CCHCR1 30
362 ENSG00000204568 6 MRPS18B 5
363 ENSG00000205138 19 SDHAF1 1
364 ENSG00000205476 14 CCDC85C 7
365 ENSG00000205572 5 SERF1B 8
366 ENSG00000205659 14 LIN52 5
367 ENSG00000205937 16 RNPS1 26
368 ENSG00000213024 19 NUP62 24
369 ENSG00000213614 15 HEXA 18
370 ENSG00000213762 19 ZNF134 4
371 ENSG00000215114 8 UBXN2B 5
372 ENSG00000221909 7 FAM200A 2
373 ENSG00000225921 6 NOL7 3
374 ENSG00000228474 2 OST4 3
375 ENSG00000229644 10 NAMPTL 2
376 ENSG00000240849 20 TMEM189 6
377 ENSG00000242612 16 DECR2 12
378 ENSG00000247626 2 MARS2 1
379 ENSG00000254858 19 MPV17L2 4
380 ENSG00000255103 1 KIAA0754 1
381 ENSG00000256525 17 POLG2 10
382 ENSG00000257315 1 ZBED6 1
383 ENSG00000261052 16 SULT1A3 13
384 ENSG00000264364 17 DYNLL2 1
385 ENSG00000269404 19 SPIB 8
description
1 mitogen-activated protein kinase kinase kinase 9 [Source:HGNC Symbol;Acc:6861]
2 proteasome (prosome, macropain) subunit, beta type, 1 [Source:HGNC Symbol;Acc:9537]
3 aminoadipate-semialdehyde synthase [Source:HGNC Symbol;Acc:17366]
4 ribosomal protein S20 [Source:HGNC Symbol;Acc:10405]
5 SprT-like N-terminal domain [Source:HGNC Symbol;Acc:25356]
6 chloride channel, voltage-sensitive 6 [Source:HGNC Symbol;Acc:2024]
7 BTB (POZ) domain containing 7 [Source:HGNC Symbol;Acc:18269]
8 ring finger protein 14 [Source:HGNC Symbol;Acc:10058]
9 rabaptin, RAB GTPase binding effector protein 1 [Source:HGNC Symbol;Acc:17677]
10 IKAROS family zinc finger 2 (Helios) [Source:HGNC Symbol;Acc:13177]
11 fucosyltransferase 8 (alpha (1,6) fucosyltransferase) [Source:HGNC Symbol;Acc:4019]
12 mitochondrial calcium uniporter regulator 1 [Source:HGNC Symbol;Acc:21097]
13 M-phase phosphoprotein 9 [Source:HGNC Symbol;Acc:7215]
14 cell division cycle and apoptosis regulator 1 [Source:HGNC Symbol;Acc:24236]
15 citrate synthase [Source:HGNC Symbol;Acc:2422]
16 ribosomal protein L18 [Source:HGNC Symbol;Acc:10310]
17 TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa [Source:HGNC Symbol;Acc:11544]
18 zinc finger CCCH-type containing 15 [Source:HGNC Symbol;Acc:29528]
19 nuclear transcription factor Y, gamma [Source:HGNC Symbol;Acc:7806]
20 GRIP1 associated protein 1 [Source:HGNC Symbol;Acc:18706]
21 phosphatidylinositol glycan anchor biosynthesis, class B [Source:HGNC Symbol;Acc:8959]
22 poly(A) binding protein, cytoplasmic 1 [Source:HGNC Symbol;Acc:8554]
23 mitogen-activated protein kinase kinase kinase kinase 4 [Source:HGNC Symbol;Acc:6866]
24 minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:6944]
25 tetratricopeptide repeat domain 38 [Source:HGNC Symbol;Acc:26082]
26 sperm associated antigen 5 [Source:HGNC Symbol;Acc:13452]
27 melanoma cell adhesion molecule [Source:HGNC Symbol;Acc:6934]
28 solute carrier family 25, member 43 [Source:HGNC Symbol;Acc:30557]
29 ubiquitin-conjugating enzyme E2D 4 (putative) [Source:HGNC Symbol;Acc:21647]
30 solute carrier family 1 (glial high affinity glutamate transporter), member 3 [Source:HGNC Symbol;Acc:10941]
31 MAP kinase interacting serine/threonine kinase 1 [Source:HGNC Symbol;Acc:7110]
32 phosphoglucomutase 1 [Source:HGNC Symbol;Acc:8905]
33 RAB21, member RAS oncogene family [Source:HGNC Symbol;Acc:18263]
34 jumonji domain containing 4 [Source:HGNC Symbol;Acc:25724]
35 cortactin [Source:HGNC Symbol;Acc:3338]
36 acyl-CoA oxidase 3, pristanoyl [Source:HGNC Symbol;Acc:121]
37 phosphatidylglycerophosphate synthase 1 [Source:HGNC Symbol;Acc:30029]
38 ALG6, alpha-1,3-glucosyltransferase [Source:HGNC Symbol;Acc:23157]
39 poly(A) polymerase alpha [Source:HGNC Symbol;Acc:14981]
40 N-acetyltransferase 14 (GCN5-related, putative) [Source:HGNC Symbol;Acc:28918]
41 oxysterol binding protein-like 8 [Source:HGNC Symbol;Acc:16396]
42 TERF1 (TRF1)-interacting nuclear factor 2 [Source:HGNC Symbol;Acc:11824]
43 enoyl CoA hydratase domain containing 1 [Source:HGNC Symbol;Acc:21489]
44 acyl-CoA thioesterase 7 [Source:HGNC Symbol;Acc:24157]
45 proprotein convertase subtilisin/kexin type 5 [Source:HGNC Symbol;Acc:8747]
46 TRIO and F-actin binding protein [Source:HGNC Symbol;Acc:17009]
47 eukaryotic translation initiation factor 3, subunit L [Source:HGNC Symbol;Acc:18138]
48 MICAL-like 1 [Source:HGNC Symbol;Acc:29804]
49 THO complex 5 [Source:HGNC Symbol;Acc:19074]
50 centrosomal protein 128kDa [Source:HGNC Symbol;Acc:20359]
51 hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) [Source:HGNC Symbol;Acc:4910]
52 ribosomal protein S6 kinase, 90kDa, polypeptide 5 [Source:HGNC Symbol;Acc:10434]
53 ER membrane protein complex subunit 9 [Source:HGNC Symbol;Acc:20273]
54 breast cancer metastasis-suppressor 1-like [Source:HGNC Symbol;Acc:20512]
55 MRG/MORF4L binding protein [Source:HGNC Symbol;Acc:15866]
56 solute carrier family 17 (vesicular nucleotide transporter), member 9 [Source:HGNC Symbol;Acc:16192]
57 Na+/K+ transporting ATPase interacting 4 [Source:HGNC Symbol;Acc:16191]
58 phospholipase C, beta 4 [Source:HGNC Symbol;Acc:9059]
59 zinc finger protein 516 [Source:HGNC Symbol;Acc:28990]
60 structural maintenance of chromosomes flexible hinge domain containing 1 [Source:HGNC Symbol;Acc:29090]
61 ATPase, class VI, type 11C [Source:HGNC Symbol;Acc:13554]
62 coiled-coil domain containing 22 [Source:HGNC Symbol;Acc:28909]
63 retinoblastoma binding protein 7 [Source:HGNC Symbol;Acc:9890]
64 solute carrier family 25 (mitochondrial carrier, brain), member 14 [Source:HGNC Symbol;Acc:10984]
65 magnesium transporter 1 [Source:HGNC Symbol;Acc:28880]
66 early endosome antigen 1 [Source:HGNC Symbol;Acc:3185]
67 ubiquitin specific peptidase 11 [Source:HGNC Symbol;Acc:12609]
68 fibronectin type III domain containing 3A [Source:HGNC Symbol;Acc:20296]
69 centromere protein T [Source:HGNC Symbol;Acc:25787]
70 crystallin, mu [Source:HGNC Symbol;Acc:2418]
71 glutamyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:29419]
72 spalt-like transcription factor 1 [Source:HGNC Symbol;Acc:10524]
73 chromosome 16 open reading frame 62 [Source:HGNC Symbol;Acc:24641]
74 transmembrane protein 87A [Source:HGNC Symbol;Acc:24522]
75 sorting nexin 16 [Source:HGNC Symbol;Acc:14980]
76 SH2 domain containing 4A [Source:HGNC Symbol;Acc:26102]
77 DOT1-like histone H3K79 methyltransferase [Source:HGNC Symbol;Acc:24948]
78 fizzy/cell division cycle 20 related 1 (Drosophila) [Source:HGNC Symbol;Acc:24824]
79 Lipid phosphate phosphatase-related protein type 2 [Source:UniProtKB/Swiss-Prot;Acc:Q96GM1]
80 neuronal pentraxin II [Source:HGNC Symbol;Acc:7953]
81 tripartite motif containing 14 [Source:HGNC Symbol;Acc:16283]
82 TBC1 domain family, member 13 [Source:HGNC Symbol;Acc:25571]
83 glutaredoxin 3 [Source:HGNC Symbol;Acc:15987]
84 DnaJ (Hsp40) homolog, subfamily C, member 12 [Source:HGNC Symbol;Acc:28908]
85 RAD51 paralog C [Source:HGNC Symbol;Acc:9820]
86 RAB5C, member RAS oncogene family [Source:HGNC Symbol;Acc:9785]
87 neuromedin U [Source:HGNC Symbol;Acc:7859]
88 aminoadipate aminotransferase [Source:HGNC Symbol;Acc:17929]
89 F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:16712]
90 stromal interaction molecule 2 [Source:HGNC Symbol;Acc:19205]
91 autophagy related 2A [Source:HGNC Symbol;Acc:29028]
92 decapping enzyme, scavenger [Source:HGNC Symbol;Acc:29812]
93 FAD-dependent oxidoreductase domain containing 1 [Source:HGNC Symbol;Acc:26927]
94 homeodomain interacting protein kinase 3 [Source:HGNC Symbol;Acc:4915]
95 MAP-kinase activating death domain [Source:HGNC Symbol;Acc:6766]
96 GLI family zinc finger 1 [Source:HGNC Symbol;Acc:4317]
97 proline rich 4 (lacrimal) [Source:HGNC Symbol;Acc:18020]
98 acyl-CoA dehydrogenase family, member 10 [Source:HGNC Symbol;Acc:21597]
99 syntaxin 2 [Source:HGNC Symbol;Acc:3403]
100 N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits [Source:HGNC Symbol;Acc:29670]
101 histidine triad nucleotide binding protein 3 [Source:HGNC Symbol;Acc:18468]
102 serine/arginine-rich splicing factor 3 [Source:HGNC Symbol;Acc:10785]
103 small ArfGAP 1 [Source:HGNC Symbol;Acc:19651]
104 GDP-mannose 4,6-dehydratase [Source:HGNC Symbol;Acc:4369]
105 vascular endothelial growth factor A [Source:HGNC Symbol;Acc:12680]
106 drebrin 1 [Source:HGNC Symbol;Acc:2695]
107 nitrogen permease regulator-like 2 (S. cerevisiae) [Source:HGNC Symbol;Acc:24969]
108 eukaryotic translation initiation factor 1B [Source:HGNC Symbol;Acc:30792]
109 phosducin-like 3 [Source:HGNC Symbol;Acc:28860]
110 inhibitor of DNA binding 2, dominant negative helix-loop-helix protein [Source:HGNC Symbol;Acc:5361]
111 regulator of microtubule dynamics 2 [Source:HGNC Symbol;Acc:26567]
112 ERBB receptor feedback inhibitor 1 [Source:HGNC Symbol;Acc:18185]
113 dedicator of cytokinesis 7 [Source:HGNC Symbol;Acc:19190]
114 solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 [Source:HGNC Symbol;Acc:20800]
115 signal-induced proliferation-associated 1 like 2 [Source:HGNC Symbol;Acc:23800]
116 v-akt murine thymoma viral oncogene homolog 3 [Source:HGNC Symbol;Acc:393]
117 RAB7, member RAS oncogene family-like 1 [Source:HGNC Symbol;Acc:9789]
118 calmodulin regulated spectrin-associated protein family, member 2 [Source:HGNC Symbol;Acc:29188]
119 F-box and leucine-rich repeat protein 5 [Source:HGNC Symbol;Acc:13602]
120 DDB1 and CUL4 associated factor 4 [Source:HGNC Symbol;Acc:20229]
121 feline leukemia virus subgroup C cellular receptor family, member 2 [Source:HGNC Symbol;Acc:20105]
122 B-cell CLL/lymphoma 11A (zinc finger protein) [Source:HGNC Symbol;Acc:13221]
123 glutamic-oxaloacetic transaminase 1, soluble [Source:HGNC Symbol;Acc:4432]
124 NudC domain containing 1 [Source:HGNC Symbol;Acc:24306]
125 plastin 1 [Source:HGNC Symbol;Acc:9090]
126 A kinase (PRKA) anchor protein 1 [Source:HGNC Symbol;Acc:367]
127 desumoylating isopeptidase 2 [Source:HGNC Symbol;Acc:24264]
128 peroxisomal biogenesis factor 16 [Source:HGNC Symbol;Acc:8857]
129 mediator complex subunit 13-like [Source:HGNC Symbol;Acc:22962]
130 emopamil binding protein-like [Source:HGNC Symbol;Acc:18061]
131 centromere protein K [Source:HGNC Symbol;Acc:29479]
132 low density lipoprotein receptor-related protein 1 [Source:HGNC Symbol;Acc:6692]
133 solute carrier family 36 (proton/amino acid symporter), member 1 [Source:HGNC Symbol;Acc:18761]
134 MAX dimerization protein 4 [Source:HGNC Symbol;Acc:13906]
135 syndecan 4 [Source:HGNC Symbol;Acc:10661]
136 glutamate receptor, metabotropic 4 [Source:HGNC Symbol;Acc:4596]
137 butyrophilin, subfamily 2, member A2 [Source:HGNC Symbol;Acc:1137]
138 SRY (sex determining region Y)-box 9 [Source:HGNC Symbol;Acc:11204]
139 armadillo repeat containing 7 [Source:HGNC Symbol;Acc:26168]
140 membrane bound O-acyltransferase domain containing 7 [Source:HGNC Symbol;Acc:15505]
141 mediator complex subunit 1 [Source:HGNC Symbol;Acc:9234]
142 centromere protein B, 80kDa [Source:HGNC Symbol;Acc:1852]
143 bone morphogenetic protein 2 [Source:HGNC Symbol;Acc:1069]
144 HECT domain containing E3 ubiquitin protein ligase 3 [Source:HGNC Symbol;Acc:26117]
145 L-3-hydroxyproline dehydratase (trans-) [Source:HGNC Symbol;Acc:20488]
146 ras homolog family member T1 [Source:HGNC Symbol;Acc:21168]
147 phosphopantothenoylcysteine synthetase [Source:HGNC Symbol;Acc:25686]
148 anaphase promoting complex subunit 13 [Source:HGNC Symbol;Acc:24540]
149 pseudouridylate synthase 7 homolog (S. cerevisiae)-like [Source:HGNC Symbol;Acc:25276]
150 solute carrier family 44 (choline transporter), member 2 [Source:HGNC Symbol;Acc:17292]
151 sorting nexin 9 [Source:HGNC Symbol;Acc:14973]
152 calmodulin regulated spectrin-associated protein 1 [Source:HGNC Symbol;Acc:19946]
153 leucine rich repeat containing 47 [Source:HGNC Symbol;Acc:29207]
154 Gse1 coiled-coil protein [Source:HGNC Symbol;Acc:28979]
155 GINS complex subunit 2 (Psf2 homolog) [Source:HGNC Symbol;Acc:24575]
156 small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:11152]
157 TBC1 domain family, member 14 [Source:HGNC Symbol;Acc:29246]
158 H3 histone, family 3B (H3.3B) [Source:HGNC Symbol;Acc:4765]
159 matrilin 2 [Source:HGNC Symbol;Acc:6908]
160 zinc finger, BED-type containing 3 [Source:HGNC Symbol;Acc:20711]
161 brain expressed, X-linked 1 [Source:HGNC Symbol;Acc:1036]
162 serine/arginine repetitive matrix 1 [Source:HGNC Symbol;Acc:16638]
163 BTB (POZ) domain containing 2 [Source:HGNC Symbol;Acc:15504]
164 CNDP dipeptidase 2 (metallopeptidase M20 family) [Source:HGNC Symbol;Acc:24437]
165 immediate early response 3 interacting protein 1 [Source:HGNC Symbol;Acc:18550]
166 tetraspanin 2 [Source:HGNC Symbol;Acc:20659]
167 prostaglandin F2 receptor inhibitor [Source:HGNC Symbol;Acc:9601]
168 Rho-associated, coiled-coil containing protein kinase 2 [Source:HGNC Symbol;Acc:10252]
169 follistatin [Source:HGNC Symbol;Acc:3971]
170 MyoD family inhibitor domain containing [Source:HGNC Symbol;Acc:28870]
171 PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:20074]
172 heterogeneous nuclear ribonucleoprotein A1 [Source:HGNC Symbol;Acc:5031]
173 basic leucine zipper and W2 domains 2 [Source:HGNC Symbol;Acc:18808]
174 membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:17393]
175 TRAF family member-associated NFKB activator [Source:HGNC Symbol;Acc:11562]
176 bridging integrator 1 [Source:HGNC Symbol;Acc:1052]
177 solute carrier family 31 (copper transporter), member 1 [Source:HGNC Symbol;Acc:11016]
178 RAN binding protein 6 [Source:HGNC Symbol;Acc:9851]
179 immediate early response 3 [Source:HGNC Symbol;Acc:5392]
180 thiopurine S-methyltransferase [Source:HGNC Symbol;Acc:12014]
181 general transcription factor IIB [Source:HGNC Symbol;Acc:4648]
182 ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) [Source:HGNC Symbol;Acc:29361]
183 ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) [Source:HGNC Symbol;Acc:167]
184 hyperpolarization activated cyclic nucleotide-gated potassium channel 4 [Source:HGNC Symbol;Acc:16882]
185 KIAA1644 [Source:HGNC Symbol;Acc:29335]
186 lysophosphatidic acid receptor 6 [Source:HGNC Symbol;Acc:15520]
187 WD repeat domain 61 [Source:HGNC Symbol;Acc:30300]
188 thioredoxin-like 4B [Source:HGNC Symbol;Acc:26041]
189 MARVEL domain containing 3 [Source:HGNC Symbol;Acc:30525]
190 glycine cleavage system protein H (aminomethyl carrier) [Source:HGNC Symbol;Acc:4208]
191 microtubule-associated protein 1 light chain 3 beta [Source:HGNC Symbol;Acc:13352]
192 ribosomal protein S2 [Source:HGNC Symbol;Acc:10404]
193 transcription factor 25 (basic helix-loop-helix) [Source:HGNC Symbol;Acc:29181]
194 growth factor receptor-bound protein 7 [Source:HGNC Symbol;Acc:4567]
195 ZFP14 zinc finger protein [Source:HGNC Symbol;Acc:29312]
196 superoxide dismutase 1, soluble [Source:HGNC Symbol;Acc:11179]
197 family with sequence similarity 71, member E1 [Source:HGNC Symbol;Acc:25107]
198 ribosomal protein L13a [Source:HGNC Symbol;Acc:10304]
199 EF-hand domain family, member D2 [Source:HGNC Symbol;Acc:28670]
200 cellular retinoic acid binding protein 2 [Source:HGNC Symbol;Acc:2339]
201 lysophospholipase-like 1 [Source:HGNC Symbol;Acc:20440]
202 CDC42 binding protein kinase alpha (DMPK-like) [Source:HGNC Symbol;Acc:1737]
203 TP53RK binding protein [Source:HGNC Symbol;Acc:24259]
204 AMMECR1-like [Source:HGNC Symbol;Acc:28658]
205 methyltransferase like 21A [Source:HGNC Symbol;Acc:30476]
206 IQ motif and Sec7 domain 1 [Source:HGNC Symbol;Acc:29112]
207 abhydrolase domain containing 10 [Source:HGNC Symbol;Acc:25656]
208 KIAA0226 [Source:HGNC Symbol;Acc:28991]
209 enolase-phosphatase 1 [Source:HGNC Symbol;Acc:24599]
210 polo-like kinase 2 [Source:HGNC Symbol;Acc:19699]
211 peptidylglycine alpha-amidating monooxygenase [Source:HGNC Symbol;Acc:8596]
212 NHP2 ribonucleoprotein [Source:HGNC Symbol;Acc:14377]
213 TBC1 domain family, member 32 [Source:HGNC Symbol;Acc:21485]
214 transmembrane protein 181 [Source:HGNC Symbol;Acc:20958]
215 ATP/GTP binding protein-like 3 [Source:HGNC Symbol;Acc:27981]
216 transmembrane protein 47 [Source:HGNC Symbol;Acc:18515]
217 zinc finger protein 185 (LIM domain) [Source:HGNC Symbol;Acc:12976]
218 ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 [Source:HGNC Symbol;Acc:854]
219 ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:12557]
220 zinc finger, CCHC domain containing 7 [Source:HGNC Symbol;Acc:26209]
221 mediator complex subunit 22 [Source:HGNC Symbol;Acc:11477]
222 REX4, RNA exonuclease 4 homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:12820]
223 CDKN1A interacting zinc finger protein 1 [Source:HGNC Symbol;Acc:16744]
224 diphthamide biosynthesis 7 [Source:HGNC Symbol;Acc:25199]
225 regulator of G-protein signaling 10 [Source:HGNC Symbol;Acc:9992]
226 serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 [Source:HGNC Symbol;Acc:1228]
227 tRNA phosphotransferase 1 [Source:HGNC Symbol;Acc:20316]
228 latrophilin 3 [Source:HGNC Symbol;Acc:20974]
229 nucleotide binding protein-like [Source:HGNC Symbol;Acc:20278]
230 geranylgeranyl diphosphate synthase 1 [Source:HGNC Symbol;Acc:4249]
231 MARVEL domain containing 2 [Source:HGNC Symbol;Acc:26401]
232 nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:21042]
233 melanoma antigen family D, 4 [Source:HGNC Symbol;Acc:23793]
234 sorbin and SH3 domain containing 2 [Source:HGNC Symbol;Acc:24098]
235 thymosin beta 4, Y-linked [Source:HGNC Symbol;Acc:11882]
236 HEAT repeat containing 3 [Source:HGNC Symbol;Acc:26087]
237 vacuolar protein sorting 37 homolog A (S. cerevisiae) [Source:HGNC Symbol;Acc:24928]
238 vacuolar protein sorting 8 homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:29122]
239 TatD DNase domain containing 2 [Source:HGNC Symbol;Acc:28988]
240 adaptor-related protein complex 3, sigma 2 subunit [Source:HGNC Symbol;Acc:571]
241 signal peptide peptidase like 3 [Source:HGNC Symbol;Acc:30424]
242 dihydropyrimidinase-like 5 [Source:HGNC Symbol;Acc:20637]
243 platelet-activating factor acetylhydrolase 2, 40kDa [Source:HGNC Symbol;Acc:8579]
244 protein tyrosine phosphatase domain containing 1 [Source:HGNC Symbol;Acc:30184]
245 NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) [Source:HGNC Symbol;Acc:7708]
246 ubiquitin-conjugating enzyme E2Z [Source:HGNC Symbol;Acc:25847]
247 ubiquitin protein ligase E3 component n-recognin 1 [Source:HGNC Symbol;Acc:16808]
248 SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:25016]
249 VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:22082]
250 ribosomal RNA processing 1 [Source:HGNC Symbol;Acc:18785]
251 TAO kinase 1 [Source:HGNC Symbol;Acc:29259]
252 SIK family kinase 3 [Source:HGNC Symbol;Acc:29165]
253 zinc finger protein 577 [Source:HGNC Symbol;Acc:28673]
254 fibroblast growth factor 11 [Source:HGNC Symbol;Acc:3667]
255 WD repeat domain 90 [Source:HGNC Symbol;Acc:26960]
256 nuclear RNA export factor 1 [Source:HGNC Symbol;Acc:8071]
257 matrix-remodelling associated 8 [Source:HGNC Symbol;Acc:7542]
258 DnaJ (Hsp40) homolog, subfamily B, member 4 [Source:HGNC Symbol;Acc:14886]
259 acid phosphatase 6, lysophosphatidic [Source:HGNC Symbol;Acc:29609]
260 frizzled-related protein [Source:HGNC Symbol;Acc:3959]
261 regulation of nuclear pre-mRNA domain containing 2 [Source:HGNC Symbol;Acc:29039]
262 chromosome 1 open reading frame 106 [Source:HGNC Symbol;Acc:25599]
263 leucine rich repeat containing 58 [Source:HGNC Symbol;Acc:26968]
264 serine/threonine kinase 36 [Source:HGNC Symbol;Acc:17209]
265 ribosomal protein L22-like 1 [Source:HGNC Symbol;Acc:27610]
266 proline-rich transmembrane protein 3 [Source:HGNC Symbol;Acc:26591]
267 BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) [Source:HGNC Symbol;Acc:950]
268 Rho guanine nucleotide exchange factor (GEF) 3 [Source:HGNC Symbol;Acc:683]
269 sprouty homolog 1, antagonist of FGF signaling (Drosophila) [Source:HGNC Symbol;Acc:11269]
270 testis expressed 264 [Source:HGNC Symbol;Acc:30247]
271 F-box protein 8 [Source:HGNC Symbol;Acc:13587]
272 ATP-binding cassette, sub-family E (OABP), member 1 [Source:HGNC Symbol;Acc:69]
273 ubiquitin specific peptidase 49 [Source:HGNC Symbol;Acc:20078]
274 somatomedin B and thrombospondin, type 1 domain containing [Source:HGNC Symbol;Acc:30362]
275 solute carrier family 25 (mitochondrial folate carrier), member 32 [Source:HGNC Symbol;Acc:29683]
276 dynein, light chain, Tctex-type 3 [Source:HGNC Symbol;Acc:11694]
277 tRNA methyltransferase 10 homolog B (S. cerevisiae) [Source:HGNC Symbol;Acc:26454]
278 pre-mRNA processing factor 18 [Source:HGNC Symbol;Acc:17351]
279 proteasome (prosome, macropain) 26S subunit, ATPase, 3 [Source:HGNC Symbol;Acc:9549]
280 cellular retinoic acid binding protein 1 [Source:HGNC Symbol;Acc:2338]
281 zinc finger protein 606 [Source:HGNC Symbol;Acc:25879]
282 signal transducer and activator of transcription 6, interleukin-4 induced [Source:HGNC Symbol;Acc:11368]
283 ubiquitin related modifier 1 [Source:HGNC Symbol;Acc:28378]
284 ENTH domain containing 2 [Source:HGNC Symbol;Acc:26458]
285 myosin VB [Source:HGNC Symbol;Acc:7603]
286 zinc finger protein 226 [Source:HGNC Symbol;Acc:13019]
287 ataxia, cerebellar, Cayman type [Source:HGNC Symbol;Acc:779]
288 Fas (TNFRSF6)-associated via death domain [Source:HGNC Symbol;Acc:3573]
289 DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A [Source:HGNC Symbol;Acc:25628]
290 ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 [Source:HGNC Symbol;Acc:12587]
291 carnitine palmitoyltransferase 1C [Source:HGNC Symbol;Acc:18540]
292 biotinidase [Source:HGNC Symbol;Acc:1122]
293 torsin A interacting protein 2 [Source:HGNC Symbol;Acc:24055]
294 transmembrane 4 L six family member 1 [Source:HGNC Symbol;Acc:11853]
295 zinc finger protein 764 [Source:HGNC Symbol;Acc:28200]
296 galactosidase, beta 1 [Source:HGNC Symbol;Acc:4298]
297 nuclear transcription factor, X-box binding-like 1 [Source:HGNC Symbol;Acc:18726]
298 KIAA0232 [Source:HGNC Symbol;Acc:28992]
299 zinc finger protein 672 [Source:HGNC Symbol;Acc:26179]
300 carbohydrate (chondroitin 4) sulfotransferase 11 [Source:HGNC Symbol;Acc:17422]
301 regulator of G-protein signaling 19 [Source:HGNC Symbol;Acc:13735]
302 RNA methyltransferase like 1 [Source:HGNC Symbol;Acc:18485]
303 Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa [Source:HGNC Symbol;Acc:7417]
304 family with sequence similarity 71, member D [Source:HGNC Symbol;Acc:20101]
305 Sjogren syndrome/scleroderma autoantigen 1 [Source:HGNC Symbol;Acc:11328]
306 FK506 binding protein 2, 13kDa [Source:HGNC Symbol;Acc:3718]
307 ArfGAP with FG repeats 1 [Source:HGNC Symbol;Acc:5175]
308 membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:28728]
309 chromosome 12 open reading frame 76 [Source:HGNC Symbol;Acc:33790]
310 transmembrane protein 167A [Source:HGNC Symbol;Acc:28330]
311 MARCKS-like 1 [Source:HGNC Symbol;Acc:7142]
312 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 [Source:HGNC Symbol;Acc:17077]
313 apoptosis-inducing, TAF9-like domain 1 [Source:HGNC Symbol;Acc:23163]
314 ras homolog family member G [Source:HGNC Symbol;Acc:672]
315 anoctamin 6 [Source:HGNC Symbol;Acc:25240]
316 ribosomal protein, large, P2 [Source:HGNC Symbol;Acc:10377]
317 adaptor-related protein complex 3, sigma 1 subunit [Source:HGNC Symbol;Acc:2013]
318 FLT3-interacting zinc finger 1 [Source:HGNC Symbol;Acc:25917]
319 dCTP pyrophosphatase 1 [Source:HGNC Symbol;Acc:28777]
320 proteasome (prosome, macropain) assembly chaperone 4 [Source:HGNC Symbol;Acc:21108]
321 regulatory factor X, 7 [Source:HGNC Symbol;Acc:25777]
322 solute carrier family 35 (GDP-fucose transporter), member C1 [Source:HGNC Symbol;Acc:20197]
323 2-aminoethanethiol (cysteamine) dioxygenase [Source:HGNC Symbol;Acc:23506]
324 chromosome 11 open reading frame 54 [Source:HGNC Symbol;Acc:30204]
325 zinc finger protein 320 [Source:HGNC Symbol;Acc:13842]
326 proline rich 14-like [Source:HGNC Symbol;Acc:28738]
327 transducer of ERBB2, 2 [Source:HGNC Symbol;Acc:11980]
328 protein tyrosine phosphatase type IVA, member 2 [Source:HGNC Symbol;Acc:9635]
329 histone cluster 1, H2ak [Source:HGNC Symbol;Acc:4726]
330 tumor necrosis factor, alpha-induced protein 2 [Source:HGNC Symbol;Acc:11895]
331 SET and MYND domain containing 3 [Source:HGNC Symbol;Acc:15513]
332 EP400 N-terminal like [Source:HGNC Symbol;Acc:26602]
333 developmentally regulated GTP binding protein 1 [Source:HGNC Symbol;Acc:3029]
334 ankyrin repeat and FYVE domain containing 1 [Source:HGNC Symbol;Acc:20763]
335 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 [Source:HGNC Symbol;Acc:16925]
336 breakpoint cluster region [Source:HGNC Symbol;Acc:1014]
337 forkhead box D3 [Source:HGNC Symbol;Acc:3804]
338 metallothionein 1X [Source:HGNC Symbol;Acc:7405]
339 prothymosin, alpha [Source:HGNC Symbol;Acc:9623]
340 uroporphyrinogen III synthase [Source:HGNC Symbol;Acc:12592]
341 S100 calcium binding protein A14 [Source:HGNC Symbol;Acc:18901]
342 histone cluster 1, H2bm [Source:HGNC Symbol;Acc:4750]
343 golgin A6 family-like 20 [Source:HGNC Symbol;Acc:49391]
344 dynein, cytoplasmic 1, heavy chain 1 [Source:HGNC Symbol;Acc:2961]
345 chromosome 20 open reading frame 112 [Source:HGNC Symbol;Acc:16106]
346 histone cluster 1, H2bh [Source:HGNC Symbol;Acc:4755]
347 myosin IC [Source:HGNC Symbol;Acc:7597]
348 transcription factor Dp-1 [Source:HGNC Symbol;Acc:11749]
349 zinc finger with KRAB and SCAN domains 8 [Source:HGNC Symbol;Acc:12983]
350 zinc finger protein 26 [Source:HGNC Symbol;Acc:13053]
351 zinc finger protein 480 [Source:HGNC Symbol;Acc:23305]
352 LPS-responsive vesicle trafficking, beach and anchor containing [Source:HGNC Symbol;Acc:1742]
353 COP9 signalosome subunit 8 [Source:HGNC Symbol;Acc:24335]
354 structural maintenance of chromosomes 5 [Source:HGNC Symbol;Acc:20465]
355 chromosome 9 open reading frame 114 [Source:HGNC Symbol;Acc:26933]
356 armadillo repeat containing, X-linked 6 [Source:HGNC Symbol;Acc:26094]
357 long intergenic non-protein coding RNA 632 [Source:HGNC Symbol;Acc:27865]
358 transcription elongation factor A (SII)-like 6 [Source:HGNC Symbol;Acc:24553]
359 negative elongation factor complex member E [Source:HGNC Symbol;Acc:13974]
360 family with sequence similarity 155, member A [Source:HGNC Symbol;Acc:33877]
361 coiled-coil alpha-helical rod protein 1 [Source:HGNC Symbol;Acc:13930]
362 mitochondrial ribosomal protein S18B [Source:HGNC Symbol;Acc:14516]
363 succinate dehydrogenase complex assembly factor 1 [Source:HGNC Symbol;Acc:33867]
364 coiled-coil domain containing 85C [Source:HGNC Symbol;Acc:35459]
365 small EDRK-rich factor 1B (centromeric) [Source:HGNC Symbol;Acc:10756]
366 lin-52 homolog (C. elegans) [Source:HGNC Symbol;Acc:19856]
367 RNA binding protein S1, serine-rich domain [Source:HGNC Symbol;Acc:10080]
368 nucleoporin 62kDa [Source:HGNC Symbol;Acc:8066]
369 hexosaminidase A (alpha polypeptide) [Source:HGNC Symbol;Acc:4878]
370 zinc finger protein 134 [Source:HGNC Symbol;Acc:12918]
371 UBX domain protein 2B [Source:HGNC Symbol;Acc:27035]
372 family with sequence similarity 200, member A [Source:HGNC Symbol;Acc:25401]
373 nucleolar protein 7, 27kDa [Source:HGNC Symbol;Acc:21040]
374 oligosaccharyltransferase 4 homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:32483]
375 nicotinamide phosphoribosyltransferase-like [Source:HGNC Symbol;Acc:17633]
376 transmembrane protein 189 [Source:HGNC Symbol;Acc:16735]
377 2,4-dienoyl CoA reductase 2, peroxisomal [Source:HGNC Symbol;Acc:2754]
378 methionyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:25133]
379 MPV17 mitochondrial membrane protein-like 2 [Source:HGNC Symbol;Acc:28177]
380 KIAA0754 [Source:HGNC Symbol;Acc:29111]
381 polymerase (DNA directed), gamma 2, accessory subunit [Source:HGNC Symbol;Acc:9180]
382 zinc finger, BED-type containing 6 [Source:HGNC Symbol;Acc:33273]
383 sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 [Source:HGNC Symbol;Acc:11455]
384 dynein, light chain, LC8-type 2 [Source:HGNC Symbol;Acc:24596]
385 Spi-B transcription factor (Spi-1/PU.1 related) [Source:HGNC Symbol;Acc:11242]Protein-protein interaction
Export significant genes.
write.table(rownames(molecules_ENSG[F.p.adj < .05, ]),
file = "../data/genelist-cv-sig.txt",
row.names = FALSE, col.names = FALSE, quote = FALSE)Use string analysis data base to look for protein protein interaction of these differential CV genes.
This is the confidence view. Stronger associations are represented by thicker lines. 
This is the evidence view. Different line colors represent the types of evidence for the association. 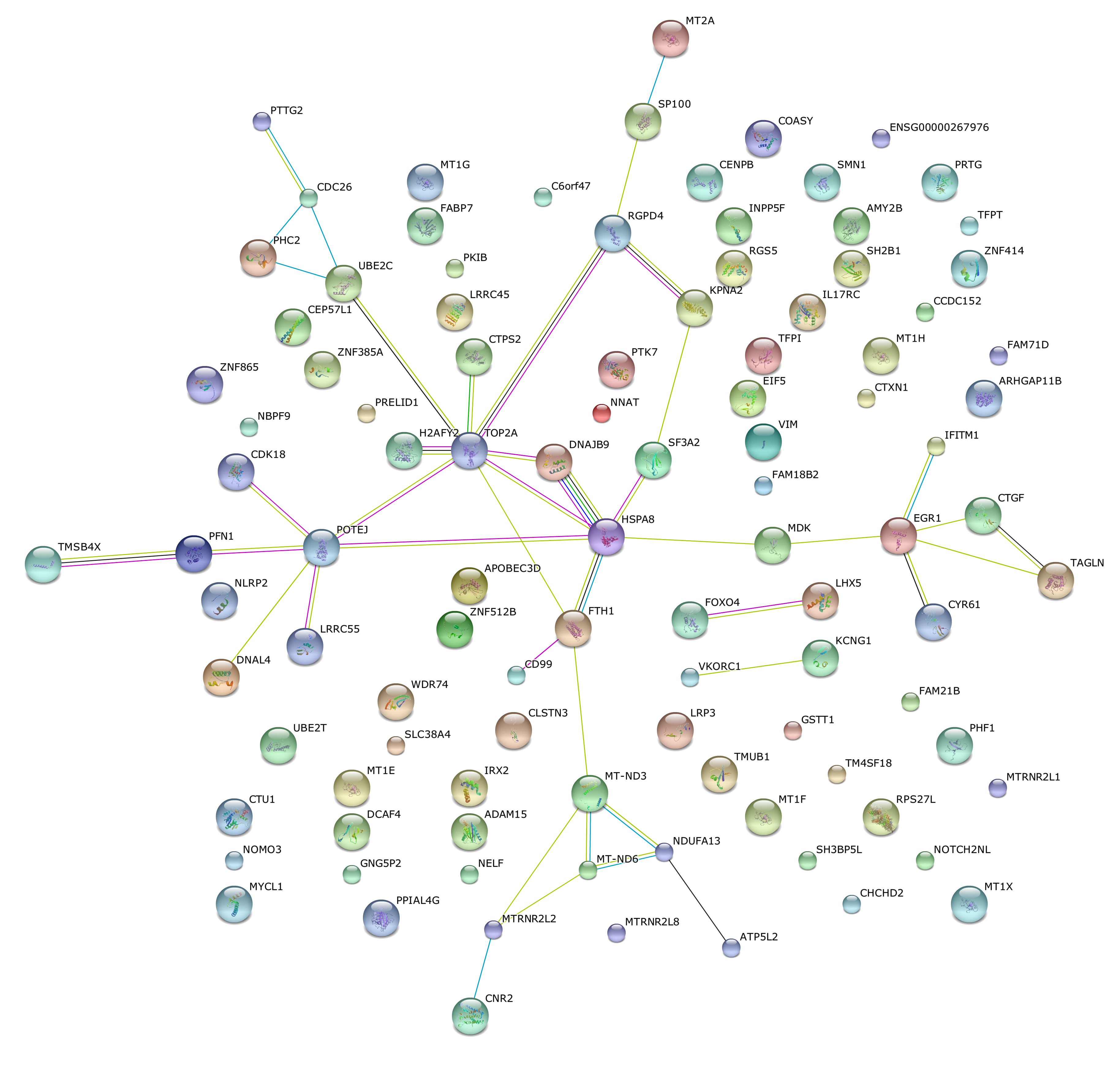
GO analysis
[Description and steps are copied from my work with Sidney]
GOstats performed Hypergeometric test for gene set enrichment. The test can take account into the conditional relationship between GO terms. Standard GOstats output contains the following information:
- GPID: GO term ID number
- Pvalue: p-vlaue of the Hypergeometric test for over-representation
- OddsRatio: effect size
- ExpCount: expected number of genes
- Count: observed number of genes
- Size: number of possible genes given the defined gene universe
GO analysis workflow is consisted of two major steps. First, we identified GO terms significant enriched given a p-value cutoff, which we set to 1 so that all possible GO terms associated with the genes can be included. We do this to eliminate false negatives. Then, we take the union of the GO terms across functional categories of interest (e.g., translation attentuation and post-translational buffering). The GO terms that are significant in at least one of the four functional categories are presented in the results.
Note. The following heatmaps are generated based on the p-values of the gene sets.
if (file.exists("rda/cv-annotation/go-cv.rda")) {
load("rda/cv-annotation/go-cv.rda")
} else {
go_sig_cv <- GOtest(my_ensembl_gene_universe = rownames(molecules_ENSG),
my_ensembl_gene_test = rownames(molecules_ENSG)[F.p.adj < .05],
pval_cutoff = 1, ontology=c("BP","CC","MF") )
save(molecules_ENSG,
F.p.adj, go_sig_cv, file = "rda/cv-annotation/go-cv.rda")
}Extract terms
if (file.exists("rda/cv-annotation/go-cv-terms.rda")) {
load("rda/cv-annotation/go-cv-terms.rda")
} else {
# Biological process
goterms_bp <- summary(go_sig_cv$GO$BP, pvalue = 1)
goterms_bp <- data.frame(ID = goterms_bp[[1]],
Pvalue = goterms_bp[[2]],
Terms = goterms_bp[[7]])
goterms_bp <- goterms_bp[order(goterms_bp$Pvalue), ]
# Cellular component
goterms_cc <- summary(go_sig_cv$GO$CC, pvalue = 1)
goterms_cc <- data.frame(ID = goterms_cc[[1]],
Pvalue = goterms_cc[[2]],
Terms = goterms_cc[[7]])
goterms_cc <- goterms_cc[order(goterms_cc$Pvalue), ]
# Molecular function
goterms_mf <- summary(go_sig_cv$GO$MF, pvalue = 1)
goterms_mf <- data.frame(ID = goterms_mf[[1]],
Pvalue = goterms_mf[[2]],
Terms = goterms_mf[[7]])
goterms_mf <- goterms_mf[order(goterms_mf$Pvalue), ]
save(goterms_bp, goterms_cc, goterms_mf,
file = "rda/cv-annotation/go-cv-terms.rda")
}*Biological process
dim(goterms_bp)[1] 1899 3kable(goterms_bp[1:50, ])| ID | Pvalue | Terms |
|---|---|---|
| GO:0071294 | 0.0000000 | cellular response to zinc ion |
| GO:0010043 | 0.0000000 | response to zinc ion |
| GO:0071276 | 0.0000000 | cellular response to cadmium ion |
| GO:0046686 | 0.0000043 | response to cadmium ion |
| GO:0071248 | 0.0000050 | cellular response to metal ion |
| GO:1990267 | 0.0000079 | response to transition metal nanoparticle |
| GO:0071241 | 0.0000120 | cellular response to inorganic substance |
| GO:0045926 | 0.0000139 | negative regulation of growth |
| GO:0010950 | 0.0001659 | positive regulation of endopeptidase activity |
| GO:0010952 | 0.0003076 | positive regulation of peptidase activity |
| GO:0036017 | 0.0004702 | response to erythropoietin |
| GO:0036018 | 0.0004702 | cellular response to erythropoietin |
| GO:0045091 | 0.0004702 | regulation of single stranded viral RNA replication via double stranded DNA intermediate |
| GO:0043280 | 0.0007251 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process |
| GO:0040008 | 0.0008488 | regulation of growth |
| GO:2001056 | 0.0009056 | positive regulation of cysteine-type endopeptidase activity |
| GO:0039692 | 0.0011619 | single stranded viral RNA replication via double stranded DNA intermediate |
| GO:0071371 | 0.0011619 | cellular response to gonadotropin stimulus |
| GO:0009611 | 0.0012159 | response to wounding |
| GO:0045862 | 0.0012961 | positive regulation of proteolysis |
| GO:0070979 | 0.0013827 | protein K11-linked ubiquitination |
| GO:0052548 | 0.0013872 | regulation of endopeptidase activity |
| GO:0070316 | 0.0016172 | regulation of G0 to G1 transition |
| GO:0010038 | 0.0017304 | response to metal ion |
| GO:0001101 | 0.0019230 | response to acid chemical |
| GO:0052547 | 0.0019253 | regulation of peptidase activity |
| GO:0045023 | 0.0021437 | G0 to G1 transition |
| GO:0071345 | 0.0033358 | cellular response to cytokine stimulus |
| GO:0034698 | 0.0034054 | response to gonadotropin |
| GO:0035456 | 0.0034054 | response to interferon-beta |
| GO:0039694 | 0.0034054 | viral RNA genome replication |
| GO:0039703 | 0.0034054 | RNA replication |
| GO:0006978 | 0.0041380 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator |
| GO:0043065 | 0.0044896 | positive regulation of apoptotic process |
| GO:0043068 | 0.0047411 | positive regulation of programmed cell death |
| GO:0042772 | 0.0049369 | DNA damage response, signal transduction resulting in transcription |
| GO:0046688 | 0.0049369 | response to copper ion |
| GO:0071229 | 0.0054035 | cellular response to acid chemical |
| GO:0048519 | 0.0056520 | negative regulation of biological process |
| GO:0060337 | 0.0057822 | type I interferon signaling pathway |
| GO:0071357 | 0.0057822 | cellular response to type I interferon |
| GO:0072210 | 0.0058007 | metanephric nephron development |
| GO:0010942 | 0.0059608 | positive regulation of cell death |
| GO:0044707 | 0.0060654 | single-multicellular organism process |
| GO:0034340 | 0.0061873 | response to type I interferon |
| GO:1903900 | 0.0065011 | regulation of viral life cycle |
| GO:0045069 | 0.0066088 | regulation of viral genome replication |
| GO:0043281 | 0.0067085 | regulation of cysteine-type endopeptidase activity involved in apoptotic process |
| GO:0001975 | 0.0067283 | response to amphetamine |
| GO:0040007 | 0.0068687 | growth |
*Cellular component
dim(goterms_cc)[1] 230 3kable(goterms_cc[1:50, ])| ID | Pvalue | Terms |
|---|---|---|
| GO:0000152 | 0.0029964 | nuclear ubiquitin ligase complex |
| GO:0032797 | 0.0045608 | SMN complex |
| GO:0097504 | 0.0045608 | Gemini of coiled bodies |
| GO:0031519 | 0.0053634 | PcG protein complex |
| GO:0019035 | 0.0094132 | viral integration complex |
| GO:0035098 | 0.0096514 | ESC/E(Z) complex |
| GO:0034719 | 0.0108718 | SMN-Sm protein complex |
| GO:0005680 | 0.0149138 | anaphase-promoting complex |
| GO:0000151 | 0.0177601 | ubiquitin ligase complex |
| GO:0008043 | 0.0187388 | intracellular ferritin complex |
| GO:0009330 | 0.0187388 | DNA topoisomerase complex (ATP-hydrolyzing) |
| GO:0070288 | 0.0187388 | ferritin complex |
| GO:0001740 | 0.0279776 | Barr body |
| GO:0097440 | 0.0279776 | apical dendrite |
| GO:0043204 | 0.0341473 | perikaryon |
| GO:0061200 | 0.0371304 | clathrin-sculpted gamma-aminobutyric acid transport vesicle |
| GO:0061202 | 0.0371304 | clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane |
| GO:0072562 | 0.0448923 | blood microparticle |
| GO:0000805 | 0.0461979 | X chromosome |
| GO:0060198 | 0.0461979 | clathrin-sculpted vesicle |
| GO:0000276 | 0.0551810 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o) |
| GO:0000974 | 0.0551810 | Prp19 complex |
| GO:0030870 | 0.0551810 | Mre11 complex |
| GO:0031461 | 0.0556633 | cullin-RING ubiquitin ligase complex |
| GO:0005681 | 0.0571076 | spliceosomal complex |
| GO:0035102 | 0.0640804 | PRC1 complex |
| GO:0015030 | 0.0642553 | Cajal body |
| GO:0048471 | 0.0781674 | perinuclear region of cytoplasm |
| GO:0000790 | 0.0868045 | nuclear chromatin |
| GO:0045263 | 0.0988565 | proton-transporting ATP synthase complex, coupling factor F(o) |
| GO:0031011 | 0.1073490 | Ino80 complex |
| GO:0033202 | 0.1073490 | DNA helicase complex |
| GO:0043025 | 0.1100471 | neuronal cell body |
| GO:0030018 | 0.1123741 | Z disc |
| GO:0008076 | 0.1157623 | voltage-gated potassium channel complex |
| GO:0005912 | 0.1219669 | adherens junction |
| GO:0034705 | 0.1240971 | potassium channel complex |
| GO:0035097 | 0.1247547 | histone methyltransferase complex |
| GO:0070161 | 0.1327431 | anchoring junction |
| GO:0031674 | 0.1342464 | I band |
| GO:0000785 | 0.1405080 | chromatin |
| GO:0031093 | 0.1486384 | platelet alpha granule lumen |
| GO:0033177 | 0.1486384 | proton-transporting two-sector ATPase complex, proton-transporting domain |
| GO:0035861 | 0.1486384 | site of double-strand break |
| GO:0034774 | 0.1566668 | secretory granule lumen |
| GO:0000228 | 0.1581837 | nuclear chromosome |
| GO:0097346 | 0.1646203 | INO80-type complex |
| GO:0044297 | 0.1691931 | cell body |
| GO:1902494 | 0.1702408 | catalytic complex |
| GO:0005925 | 0.1704578 | focal adhesion |
*Molecular function
dim(goterms_mf)[1] 285 3kable(goterms_mf[1:50, ])| ID | Pvalue | Terms |
|---|---|---|
| GO:0005520 | 0.0041803 | insulin-like growth factor binding |
| GO:0004556 | 0.0082278 | alpha-amylase activity |
| GO:0004595 | 0.0082278 | pantetheine-phosphate adenylyltransferase activity |
| GO:0004949 | 0.0082278 | cannabinoid receptor activity |
| GO:0016160 | 0.0082278 | amylase activity |
| GO:0008270 | 0.0089991 | zinc ion binding |
| GO:0061631 | 0.0093864 | ubiquitin conjugating enzyme activity |
| GO:0061650 | 0.0104352 | ubiquitin-like protein conjugating enzyme activity |
| GO:0046914 | 0.0112368 | transition metal ion binding |
| GO:0008201 | 0.0119451 | heparin binding |
| GO:0005178 | 0.0130937 | integrin binding |
| GO:0003883 | 0.0163887 | CTP synthase activity |
| GO:0004140 | 0.0163887 | dephospho-CoA kinase activity |
| GO:0016900 | 0.0163887 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor |
| GO:0032090 | 0.0163887 | Pyrin domain binding |
| GO:0047057 | 0.0163887 | vitamin-K-epoxide reductase (warfarin-sensitive) activity |
| GO:1990254 | 0.0163887 | keratin filament binding |
| GO:0005539 | 0.0220635 | glycosaminoglycan binding |
| GO:0004322 | 0.0244834 | ferroxidase activity |
| GO:0005251 | 0.0244834 | delayed rectifier potassium channel activity |
| GO:0008199 | 0.0244834 | ferric iron binding |
| GO:0016724 | 0.0244834 | oxidoreductase activity, oxidizing metal ions, oxygen as acceptor |
| GO:1990050 | 0.0244834 | phosphatidic acid transporter activity |
| GO:0000774 | 0.0325124 | adenyl-nucleotide exchange factor activity |
| GO:0003918 | 0.0325124 | DNA topoisomerase type II (ATP-hydrolyzing) activity |
| GO:0005212 | 0.0325124 | structural constituent of eye lens |
| GO:0061505 | 0.0325124 | DNA topoisomerase II activity |
| GO:0003696 | 0.0404761 | satellite DNA binding |
| GO:0008494 | 0.0404761 | translation activator activity |
| GO:0055131 | 0.0404761 | C3HC4-type RING finger domain binding |
| GO:0003916 | 0.0483751 | DNA topoisomerase activity |
| GO:0019215 | 0.0483751 | intermediate filament binding |
| GO:0030368 | 0.0483751 | interleukin-17 receptor activity |
| GO:0070087 | 0.0483751 | chromo shadow domain binding |
| GO:0008083 | 0.0485652 | growth factor activity |
| GO:0050839 | 0.0540124 | cell adhesion molecule binding |
| GO:0044877 | 0.0550545 | macromolecular complex binding |
| GO:0019237 | 0.0562100 | centromeric DNA binding |
| GO:1901681 | 0.0605070 | sulfur compound binding |
| GO:0008144 | 0.0635227 | drug binding |
| GO:0004862 | 0.0639812 | cAMP-dependent protein kinase inhibitor activity |
| GO:0008139 | 0.0639812 | nuclear localization sequence binding |
| GO:0042826 | 0.0680492 | histone deacetylase binding |
| GO:0008301 | 0.0716892 | DNA binding, bending |
| GO:0090079 | 0.0716892 | translation regulator activity, nucleic acid binding |
| GO:0048038 | 0.0793346 | quinone binding |
| GO:0051787 | 0.0793346 | misfolded protein binding |
| GO:0061630 | 0.0846794 | ubiquitin protein ligase activity |
| GO:0008656 | 0.0869178 | cysteine-type endopeptidase activator activity involved in apoptotic process |
| GO:0016722 | 0.0869178 | oxidoreductase activity, oxidizing metal ions |
Session information
sessionInfo()R version 3.2.1 (2015-06-18)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X 10.10.5 (Yosemite)
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] biomaRt_2.24.1 gridExtra_2.0.0 broman_0.59-5 zoo_1.7-12
[5] ggplot2_1.0.1 edgeR_3.10.5 limma_3.24.15 dplyr_0.4.3
[9] data.table_1.9.6 knitr_1.11
loaded via a namespace (and not attached):
[1] Rcpp_0.12.2 highr_0.5.1 GenomeInfoDb_1.4.3
[4] formatR_1.2.1 plyr_1.8.3 bitops_1.0-6
[7] tools_3.2.1 digest_0.6.8 RSQLite_1.0.0
[10] evaluate_0.8 gtable_0.1.2 lattice_0.20-33
[13] DBI_0.3.1 yaml_2.1.13 parallel_3.2.1
[16] proto_0.3-10 stringr_1.0.0 IRanges_2.2.9
[19] S4Vectors_0.6.6 stats4_3.2.1 Biobase_2.28.0
[22] R6_2.1.1 AnnotationDbi_1.30.1 XML_3.98-1.3
[25] rmarkdown_0.8.1 reshape2_1.4.1 magrittr_1.5
[28] BiocGenerics_0.14.0 scales_0.3.0 htmltools_0.2.6
[31] MASS_7.3-45 assertthat_0.1 colorspace_1.2-6
[34] labeling_0.3 stringi_1.0-1 RCurl_1.95-4.7
[37] munsell_0.4.2 chron_2.3-47