Data transformation
2015-09-30
Last updated: 2016-11-08
Code version: bd286a36f14d3b332285cdc7e62258b1f616bb14
library("ggplot2")
library("cowplot")
theme_set(theme_bw(base_size = 16))
theme_update(panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.major.y = element_blank(),
legend.key = element_blank(),
plot.title = element_text(size = rel(1)))
source("functions.R")This file performs principal components analysis (PCA) and displays the results for the data at each stage of our data transformation pipeline. Furthermore, it quantifies and tests the PCA results using pca-utils (commit 8540ff2). The methodology is described in Worley et al., 2013. Briefly, the differences between samples are calculated using the squared Mahalanobis distance. The distances are then scaled so that the hypothesis test that the samples from two groups come from separate distributions can be computed with an F-test.
Input
Input filtered annotation.
anno_filter <- read.table("../data/annotation-filter.txt", header = TRUE,
stringsAsFactors = FALSE)
head(anno_filter) individual replicate well batch sample_id
1 NA19098 r1 A01 NA19098.r1 NA19098.r1.A01
2 NA19098 r1 A02 NA19098.r1 NA19098.r1.A02
3 NA19098 r1 A04 NA19098.r1 NA19098.r1.A04
4 NA19098 r1 A05 NA19098.r1 NA19098.r1.A05
5 NA19098 r1 A06 NA19098.r1 NA19098.r1.A06
6 NA19098 r1 A07 NA19098.r1 NA19098.r1.A07Input filtered molecule counts.
molecules_filter <- read.table("../data/molecules-filter.txt", header = TRUE,
stringsAsFactors = FALSE)
stopifnot(ncol(molecules_filter) == nrow(anno_filter),
colnames(molecules_filter) == anno_filter$sample_id)Input standardized molecule counts.
molecules_cpm <- read.table("../data/molecules-cpm.txt", header = TRUE,
stringsAsFactors = FALSE)
stopifnot(ncol(molecules_cpm) == nrow(anno_filter),
colnames(molecules_cpm) == anno_filter$sample_id)Input Poisson GLM transformed molecule counts per million.
molecules_cpm_trans <- read.table("../data/molecules-cpm-trans.txt", header = TRUE,
stringsAsFactors = FALSE)
stopifnot(ncol(molecules_cpm_trans) == nrow(anno_filter),
colnames(molecules_cpm_trans) == anno_filter$sample_id)Input final batch-corrected molecule counts per million.
molecules_final <- read.table("../data/molecules-final.txt", header = TRUE,
stringsAsFactors = FALSE)
stopifnot(ncol(molecules_final) == nrow(anno_filter),
colnames(molecules_final) == anno_filter$sample_id)PCA
PCA of filtered data
pca_molecules_filter <- run_pca(molecules_filter)
pca_molecules_filter_title <- "The effect of technical batch on single cell gene expression for raw counts data"
pca_molecules_filter_plot <- plot_pca(pca_molecules_filter$PCs,
explained = pca_molecules_filter$explained,
metadata = anno_filter, color = "individual",
shape = "replicate", alpha = 0.5, size = 3) +
# labs(title = "The effect of technical batch \n on single cell gene expression data \n when raw counts are used")
labs(title = paste(strwrap(pca_molecules_filter_title, width = 50), collapse = "\n"))pca_molecules_filter_out <- data.frame(Obs.id = 1:nrow(anno_filter),
Obs.batch = anno_filter$batch,
pca_molecules_filter$PCs[, 1:2])
write.table(pca_molecules_filter_out,
file = "../data/pca-molecules-filter.txt",
quote = FALSE, sep = "\t", row.names = FALSE)pca-distances -i ../data/pca-molecules-filter.txt NA19098.r3 NA19101.r1 NA19101.r2 NA19101.r3 NA19239.r1 NA19239.r2 NA19239.r3
NA19098.r1 3.533561e+01 2.795329e+01 3.108176e+01 3.685457e+01 3.662411e+01 3.119577e+01 4.383286e+01
NA19098.r3 6.325114e+01 6.619283e+01 7.215802e+01 6.176294e+01 6.277506e+01 6.288494e+01
NA19101.r1 7.570174e+00 8.906888e+00 3.210637e+01 1.794815e+01 4.380363e+01
NA19101.r2 9.531213e+00 3.967157e+01 2.543646e+01 5.137277e+01
NA19101.r3 3.581286e+01 2.135155e+01 4.770952e+01
NA19239.r1 1.452473e+01 1.189914e+01
NA19239.r2 2.641617e+01pca-dendrogram -i ../data/pca-molecules-filter.txt
+-----NA19101.r1
+----------------------|3.5e-03
| +-------NA19101.r3
+--------------------------------------|0
| | +--------------------NA19098.r3
| | |0
| +--------------| +-------NA19098.r1
|0 +---------|6.9e-07
| +--------NA19101.r2
|
| +--------------------------------------NA19239.r2
| |0
+-----------------------------------------------------------| +-------------------NA19239.r1
+-----------------|0
+------------------NA19239.r3
PCA of standardized data
pca_molecules_cpm <- run_pca(molecules_cpm)
pca_molecules_cpm_title <- "The effect of technical batch on single cell gene expression for log counts per million data"
pca_molecules_cpm_plot <- plot_pca(pca_molecules_cpm$PCs,
explained = pca_molecules_cpm$explained,
metadata = anno_filter, color = "individual",
shape = "replicate", alpha = 0.5, size = 3) +
labs(title = paste(strwrap(pca_molecules_cpm_title, width = 50), collapse = "\n"))
# labs(title = "The effect of technical batch \n on single cell gene expression data \n when log transformed counts per million data are used")pca_molecules_cpm_out <- data.frame(Obs.id = 1:nrow(anno_filter),
Obs.batch = anno_filter$batch,
pca_molecules_cpm$PCs[, 1:2])
write.table(pca_molecules_cpm_out,
file = "../data/pca-molecules-cpm.txt",
quote = FALSE, sep = "\t", row.names = FALSE)pca-distances -i ../data/pca-molecules-cpm.txt NA19098.r3 NA19101.r1 NA19101.r2 NA19101.r3 NA19239.r1 NA19239.r2 NA19239.r3
NA19098.r1 2.283127e+01 1.934100e+01 5.296175e+01 2.764144e+01 4.969837e+01 3.699837e+01 4.594807e+01
NA19098.r3 5.978405e+00 3.026961e+01 8.082123e+00 3.866189e+01 2.588141e+01 4.246380e+01
NA19101.r1 3.508682e+01 8.325224e+00 3.575672e+01 2.257645e+01 3.777460e+01
NA19101.r2 2.848375e+01 4.631674e+01 3.973253e+01 5.806423e+01
NA19101.r3 3.087511e+01 1.850056e+01 3.614705e+01
NA19239.r1 1.323738e+01 1.539228e+01
NA19239.r2 1.869540e+01pca-dendrogram -i ../data/pca-molecules-cpm.txt
+-------------------------------NA19239.r1
+-----------------------------------------------------------|0
| | +-------------------NA19239.r2
| +----------------|0
|0 +-----------------NA19239.r3
|
| +--------------------------------------------------NA19098.r1
| |0
+--------------------------------------| +----------------------------------------NA19101.r2
+-----------------|0
| +-------------------NA19101.r3
+----------------|0
| +------NA19098.r3
+--------|7.2e-10
+-----NA19101.r1
PCA of Poisson GLM transformed molecule counts per million
pca_molecules_cpm_trans <- run_pca(molecules_cpm_trans)
pca_molecules_cpm_trans_title <- "The effect of technical batch on single cell gene expression for Poisson transformed data"
pca_molecules_cpm_trans_plot <- plot_pca(pca_molecules_cpm_trans$PCs,
explained = pca_molecules_cpm_trans$explained,
metadata = anno_filter, color = "individual",
shape = "replicate", alpha = 0.5, size = 3) +
labs(title = paste(strwrap(pca_molecules_cpm_trans_title, width = 50), collapse = "\n"))
# labs(title = "The effect of technical batch \n on single cell gene expression data \n when Poisson transformed data are used")pca_molecules_cpm_trans_out <- data.frame(Obs.id = 1:nrow(anno_filter),
Obs.batch = anno_filter$batch,
pca_molecules_cpm_trans$PCs[, 1:2])
write.table(pca_molecules_cpm_trans_out,
file = "../data/pca-molecules-cpm-trans.txt",
quote = FALSE, sep = "\t", row.names = FALSE)pca-distances -i ../data/pca-molecules-cpm-trans.txt NA19098.r3 NA19101.r1 NA19101.r2 NA19101.r3 NA19239.r1 NA19239.r2 NA19239.r3
NA19098.r1 1.656283e+01 6.146432e+01 1.321773e+01 4.332598e+01 4.827787e+01 4.750276e+01 4.949364e+01
NA19098.r3 7.802604e+01 2.497308e+01 5.987233e+01 6.155863e+01 6.265747e+01 6.149072e+01
NA19101.r1 5.731654e+01 1.845042e+01 4.202117e+01 2.987954e+01 4.863510e+01
NA19101.r2 4.062958e+01 5.399017e+01 5.019805e+01 5.661237e+01
NA19101.r3 3.109027e+01 1.973393e+01 3.732732e+01
NA19239.r1 1.225009e+01 6.618308e+00
NA19239.r2 1.885694e+01pca-dendrogram -i ../data/pca-molecules-cpm-trans.txt
+---------------------------------NA19239.r2
|0
+----------------------------------------------------------| +--------NA19239.r1
| +-------------|0
| +-------NA19239.r3
|0
| +------NA19101.r1
| +------------------------------------|0
| | +-----------NA19101.r3
+------------------------------------|0
| +------------------NA19101.r2
| |0
+---------------------| +-----NA19098.r1
+-------|0
+---------NA19098.r3
PCA of final batch-corrected data
pca_final <- run_pca(molecules_final)
pca_final_title <- "PCA of standardized, normalized, and batch corrected single cell gene expression data"
pca_final_plot <- plot_pca(pca_final$PCs, explained = pca_final$explained,
metadata = anno_filter, color = "individual",
shape = "replicate", alpha = 0.5, size = 3) +
labs(title = paste(strwrap(pca_final_title, width = 50), collapse = "\n"))
# labs(title = "PCA of standardized, \n normalized, and batch corrected \n single cell gene expression data")pca_final_out <- data.frame(Obs.id = 1:nrow(anno_filter),
Obs.batch = anno_filter$batch,
pca_final$PCs[, 1:2])
write.table(pca_final_out,
file = "../data/pca-final.txt",
quote = FALSE, sep = "\t", row.names = FALSE)pca-distances -i ../data/pca-final.txt NA19098.r3 NA19101.r1 NA19101.r2 NA19101.r3 NA19239.r1 NA19239.r2 NA19239.r3
NA19098.r1 5.080952e+00 5.435574e+01 3.835040e+01 4.975047e+01 5.613992e+01 5.478609e+01 5.546367e+01
NA19098.r3 5.926557e+01 4.294858e+01 5.468422e+01 6.085990e+01 5.961170e+01 6.014592e+01
NA19101.r1 1.830388e+01 4.708103e+00 3.624496e+01 3.249704e+01 3.706498e+01
NA19101.r2 1.472089e+01 4.267492e+01 3.927972e+01 4.297114e+01
NA19101.r3 3.429283e+01 3.056554e+01 3.498175e+01
NA19239.r1 3.748794e+00 1.333070e+00
NA19239.r2 4.658342e+00pca-dendrogram -i ../data/pca-final.txt
+------------NA19239.r2
|2.4e-08
+-----------------------------------------------------------| +-----NA19239.r1
| +-------|0.32
| +-----NA19239.r3
|0
| +------NA19098.r1
| +-------------------------------------|0.02
| | +-------NA19098.r3
+---------------------------------------|0
| +--------------NA19101.r2
| |4.3e-11
+---------------------------| +------NA19101.r1
+---------|7.7e-03
+--------NA19101.r3
Data transformation figure
plot_final <- plot_grid(
pca_molecules_filter_plot +
scale_color_discrete(name = "Individual") +
guides(shape = FALSE) + theme(legend.position = "bottom"),
pca_molecules_cpm_plot +
scale_shape(name = "Replicate") +
guides(color = FALSE) + theme(legend.position = "bottom"),
pca_molecules_cpm_trans_plot +
scale_color_discrete(name = "Individual") +
guides(shape = FALSE) + theme(legend.position = "bottom"),
pca_final_plot +
scale_shape(name = "Replicate") +
guides(color = FALSE) + theme(legend.position = "bottom"),
labels = letters[1:4])
plot_final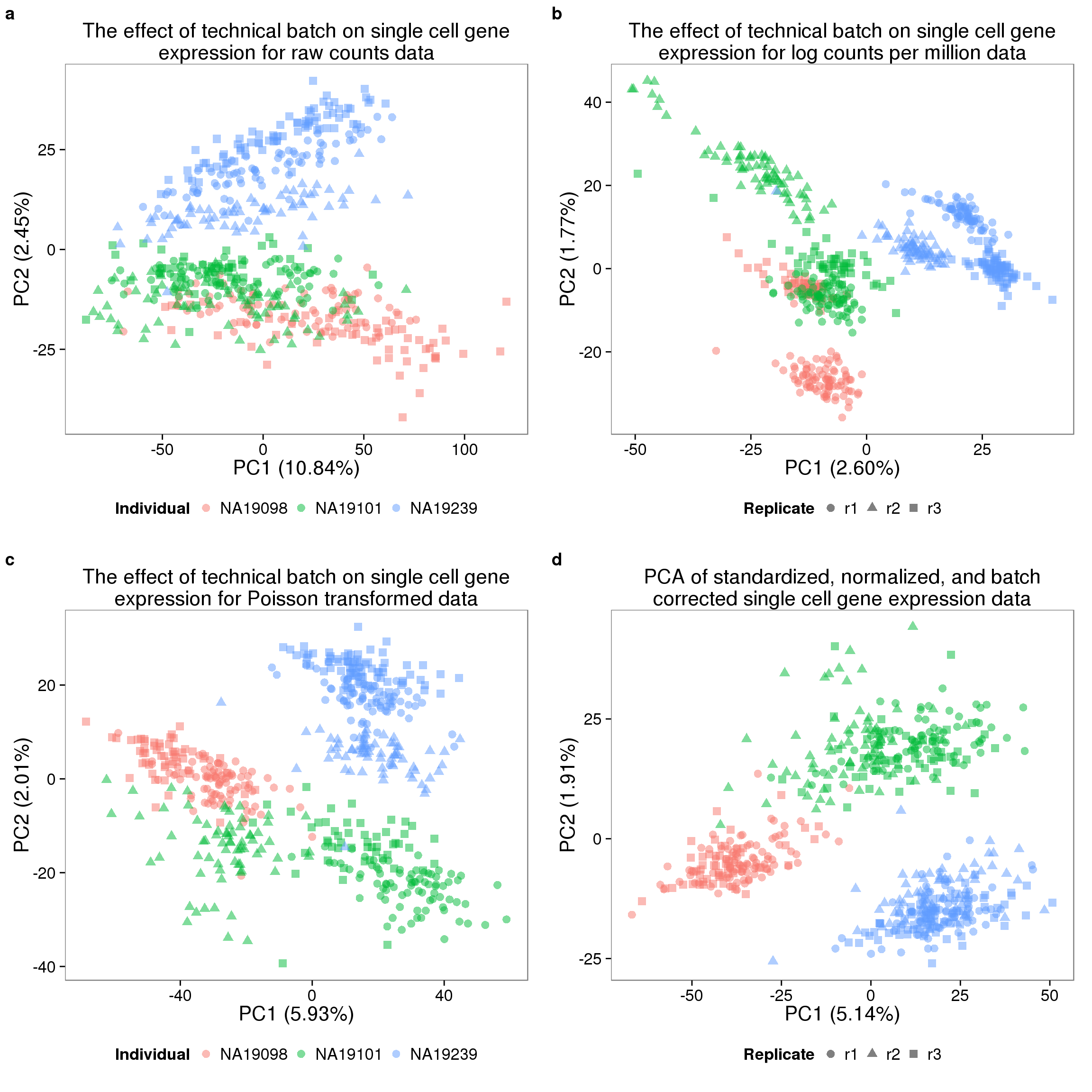
tiff("../paper/figure/fig-data-transformation.tiff",
width = 12, height = 12,
units = "in", res = 300, compression = "zip")
plot_final
dev.off()png
2 Session information
sessionInfo()R version 3.2.0 (2015-04-16)
Platform: x86_64-unknown-linux-gnu (64-bit)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] testit_0.4 cowplot_0.3.1 ggplot2_1.0.1 knitr_1.10.5
loaded via a namespace (and not attached):
[1] Rcpp_0.12.4 magrittr_1.5 MASS_7.3-40 munsell_0.4.3
[5] colorspace_1.2-6 stringr_1.0.0 httr_0.6.1 plyr_1.8.3
[9] tools_3.2.0 grid_3.2.0 gtable_0.1.2 htmltools_0.2.6
[13] yaml_2.1.13 digest_0.6.8 reshape2_1.4.1 formatR_1.2
[17] bitops_1.0-6 RCurl_1.95-4.6 evaluate_0.7 rmarkdown_0.6.1
[21] labeling_0.3 stringi_1.0-1 scales_0.4.0 proto_0.3-10