Proportion of genes detected using all cells
2016-02-16
Last updated: 2016-02-16
Code version: 19e0b5238d17b956c12a2c43d67bdc024117580c
source("functions.R")
library("edgeR")
library("tidyr")
library("ggplot2")
library("cowplot")
theme_set(theme_bw(base_size = 16))
theme_update(panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.major.y = element_blank())Hicks et al., 2015 observed a strong relationship between the proportion of genes detected (PGD) per sample and PC1 across multiple single cell studies. Here we investigate this effect in our study. Specifcally, we explore the following questions:
- How much is the PGD effect driven by lowly expressed genes?
- Does our normalization procedure reduce the effect of the PGD?
- Is there a difference in PGD between reads and molecules?
- How much of an effect does quality filtering cells have?
This analysis is very similar to More exploring of the proportion of genes detected. The main difference is this analysis includes all the single cell samples.
Input
Input annotation
anno <- read.table("../data/annotation.txt", header = TRUE,
stringsAsFactors = FALSE)Input molecule counts
molecules <- read.table("../data/molecules.txt", header = TRUE,
stringsAsFactors = FALSE)Input read counts
reads <- read.table("../data/reads.txt", header = TRUE,
stringsAsFactors = FALSE)Prepare data
Some genes have zero molecule counts across the single cells. We remove these.
zero_genes <- rowSums(molecules) == 0
molecules <- molecules[!zero_genes, ]
reads <- reads[!zero_genes, ]
stopifnot(rownames(molecules) == rownames(reads))Lastly, we include a subset of ubiquitous genes which were detected in every cell, i.e. the PGD will be 1 for all samples.
ubiquitous_genes <- rowSums(molecules > 0) == ncol(molecules)
molecules_ubiq <- molecules[ubiquitous_genes, ]
reads_ubiq <- reads[ubiquitous_genes, ]Calculate PGD
Define a function to calculate PGD.
calc_pgd <- function(x) {
# Calculate the proportion of genes detected (PGD) per sample/column
#
# x - gene expression counts
#
# Returns a numeric vector
stopifnot(!is.null(dim(x)),
x == as.integer(as.matrix(x)))
num_genes_detected <- colSums(x > 0)
num_total_genes <- nrow(x)
prop_genes_detected <- num_genes_detected / num_total_genes
stopifnot(is.numeric(prop_genes_detected),
length(prop_genes_detected) == ncol(x),
prop_genes_detected <= 1,
prop_genes_detected >= 0)
return(prop_genes_detected)
}Calculate the PGD using all genes with at least one read detected in at least one single cell.
pgd_all <- calc_pgd(reads)
# pgd_filter <- calc_pgd(reads_filter)
# pgd_ubiq <- calc_pgd(reads_ubiq)
# stopifnot(pgd_ubiq == 1)
# stopifnot(pgd_all == calc_pgd(molecules))PCA - counts
Unstandardized read counts from all genes with at least one observed read.
pca <- run_pca(reads)
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "all", processing = "counts")
d_full <- dUnstandardized molecule counts from all genes with at least one observed read.
pca <- run_pca(molecules)
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "all", processing = "counts")
d_full <- rbind(d_full, d)Unstandardized read counts from set of ubiquitous genes.
pca <- run_pca(reads_ubiq)
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "ubiq", processing = "counts")
d_full <- rbind(d_full, d)Unstandardized molecule counts from set of ubiquitous genes.
pca <- run_pca(molecules_ubiq)
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "ubiq", processing = "counts")
d_full <- rbind(d_full, d)PCA - cpm
Read counts per million from all genes with at least one observed read.
pca <- run_pca(cpm(reads))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "all", processing = "cpm")
d_full <- rbind(d_full, d)Molecule counts per million from all genes with at least one observed read.
pca <- run_pca(cpm(molecules))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "all", processing = "cpm")
d_full <- rbind(d_full, d)Read counts per million from set of ubiquitous genes.
pca <- run_pca(cpm(reads_ubiq))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "ubiq", processing = "cpm")
d_full <- rbind(d_full, d)Molecule counts per million from set of ubiquitous genes.
pca <- run_pca(cpm(molecules_ubiq))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "ubiq", processing = "cpm")
d_full <- rbind(d_full, d)PCA - log2 cpm edgeR
Read log2 counts per million (edgeR) from all genes with at least one observed read.
pca <- run_pca(cpm(reads, log = TRUE))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "all", processing = "log2cpm-edgeR")
d_full <- rbind(d_full, d)Molecule log2 counts per million (edgeR) from all genes with at least one observed read.
pca <- run_pca(cpm(molecules, log = TRUE))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "all", processing = "log2cpm-edgeR")
d_full <- rbind(d_full, d)Read log2 counts per million (edgeR) from set of ubiquitous genes.
pca <- run_pca(cpm(reads_ubiq, log = TRUE))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "ubiq", processing = "log2cpm-edgeR")
d_full <- rbind(d_full, d)Molecule log2 counts per million (edgeR) from set of ubiquitous genes.
pca <- run_pca(cpm(molecules_ubiq, log = TRUE))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "ubiq", processing = "log2cpm-edgeR")
d_full <- rbind(d_full, d)PCA - log2 cpm Rafa
Read counts per million from all genes with at least one observed read.
pca <- run_pca(log2(cpm(reads) + 1))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "all", processing = "log2cpm-Rafa")
d_full <- rbind(d_full, d)Molecule counts per million from all genes with at least one observed read.
pca <- run_pca(log2(cpm(molecules) + 1))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "all", processing = "log2cpm-Rafa")
d_full <- rbind(d_full, d)Read counts per million from set of ubiquitous genes.
pca <- run_pca(log2(cpm(reads_ubiq) + 1))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "reads", genes = "ubiq", processing = "log2cpm-Rafa")
d_full <- rbind(d_full, d)Molecule counts per million from set of ubiquitous genes.
pca <- run_pca(log2(cpm(molecules_ubiq) + 1))
d <- data.frame(id = rownames(pca$PCs), pca$PCs[, 1:2],
pgd = pgd_all,
type = "molecules", genes = "ubiq", processing = "log2cpm-Rafa")
d_full <- rbind(d_full, d)Results
For the plots below, the x-axis is always the proportion of genes detected (PGD) that was calculated when considering all genes with at least one read in at least one single cell. PC1 on the y-axis is from the PCA performed on the indicated subset of genes. Note: The direction of the correlation is arbitrary because the direction of PC1 is arbitrarily assigned during PCA.
d_full <- separate(d_full, col = id, into = c("individual", "replicate", "well"),
sep = "\\.")Effect of processing on reads using all genes:
p_reads_all <- ggplot(d_full[d_full$type == "reads" & d_full$genes == "all", ],
aes(x = pgd, y = PC1)) +
geom_point(aes(color = individual), alpha = 0.5) +
geom_smooth(method = "lm") +
facet_wrap(~processing) +
theme(legend.position = "none")
p_reads_all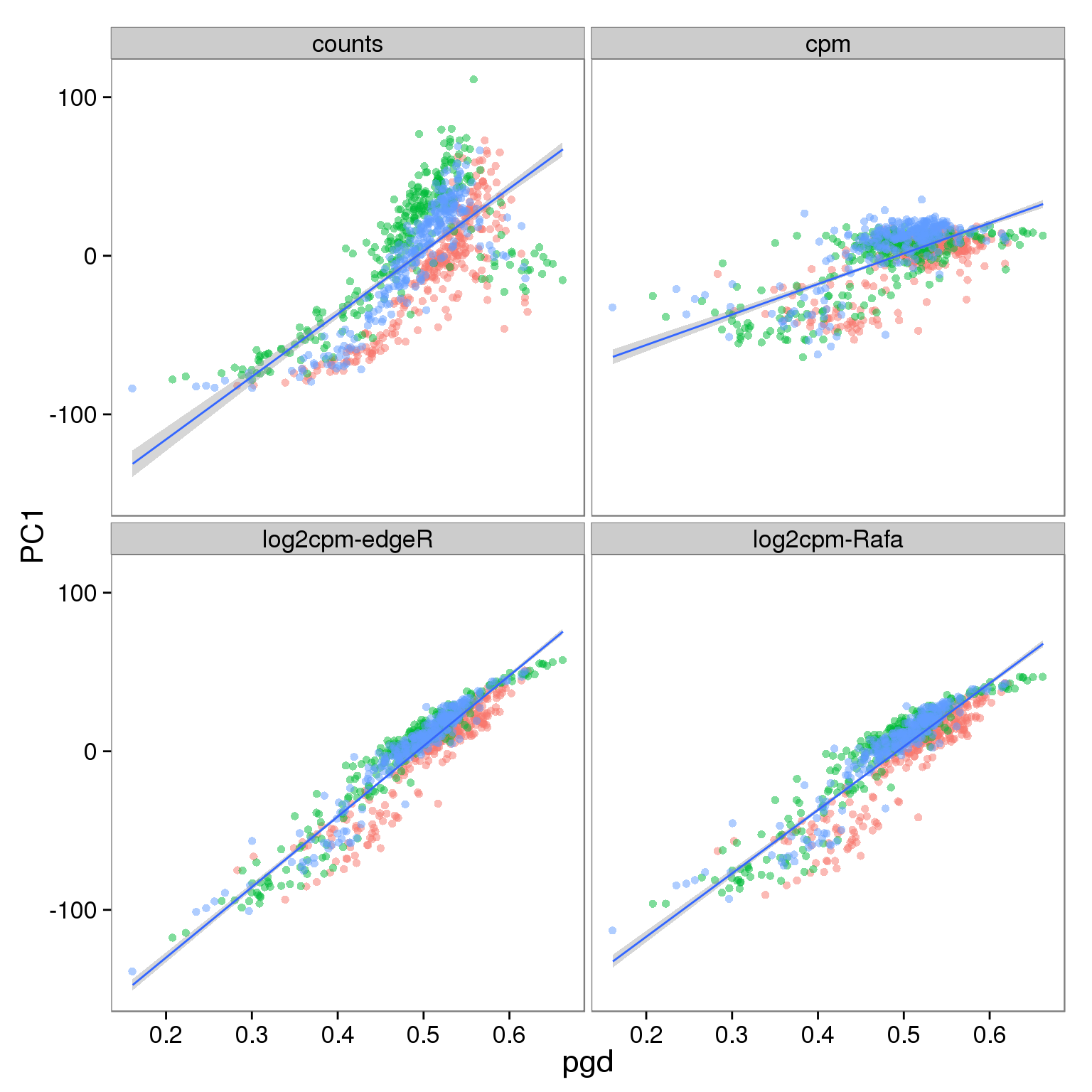
Effect of processing on molecules using all genes:
p_molecules_all <- p_reads_all %+% d_full[d_full$type == "molecules" & d_full$genes == "all", ]
p_molecules_all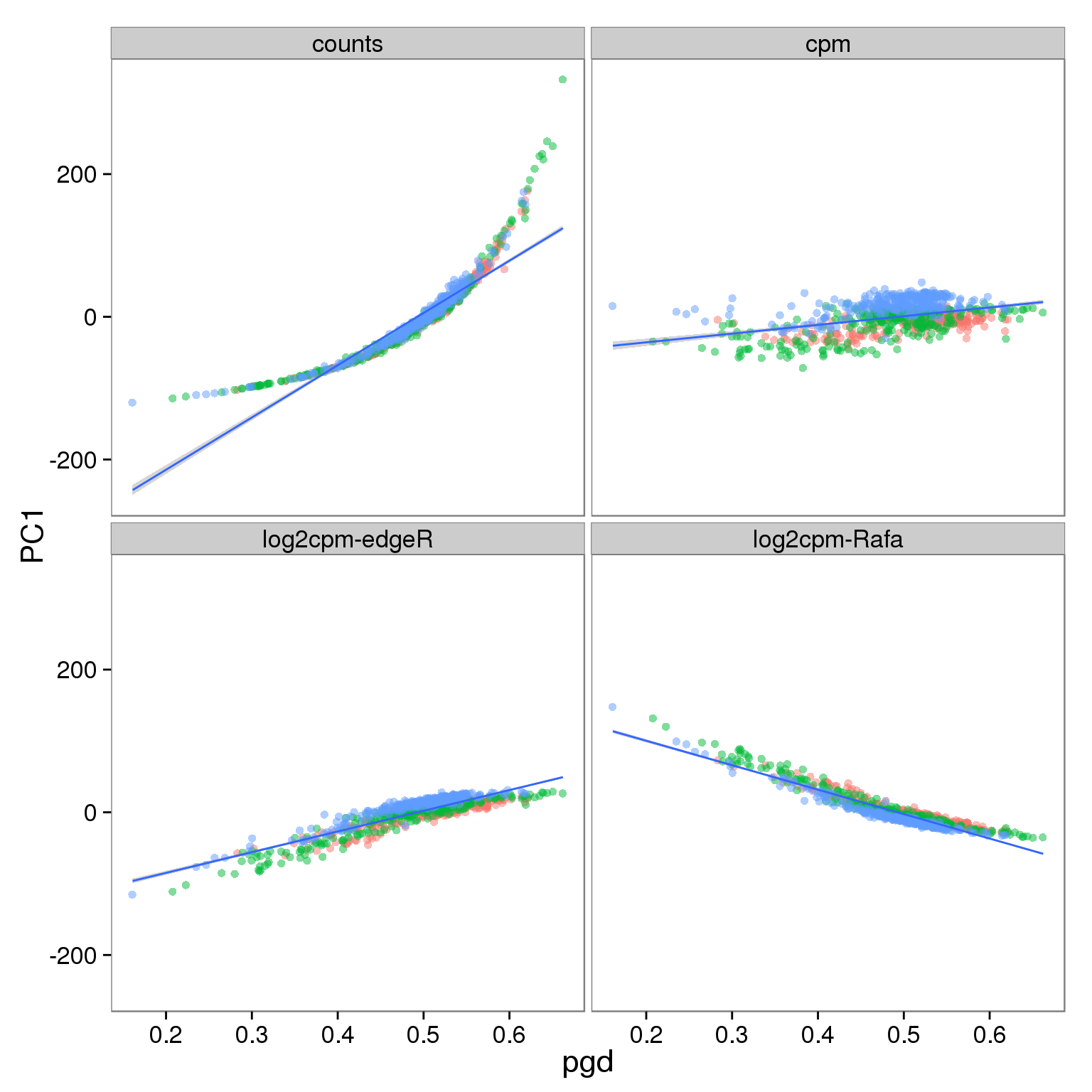
Effect of processing on reads using ubiquitous genes:
p_reads_filter <- p_reads_all %+% d_full[d_full$type == "reads" & d_full$genes == "ubiq", ]
p_reads_filter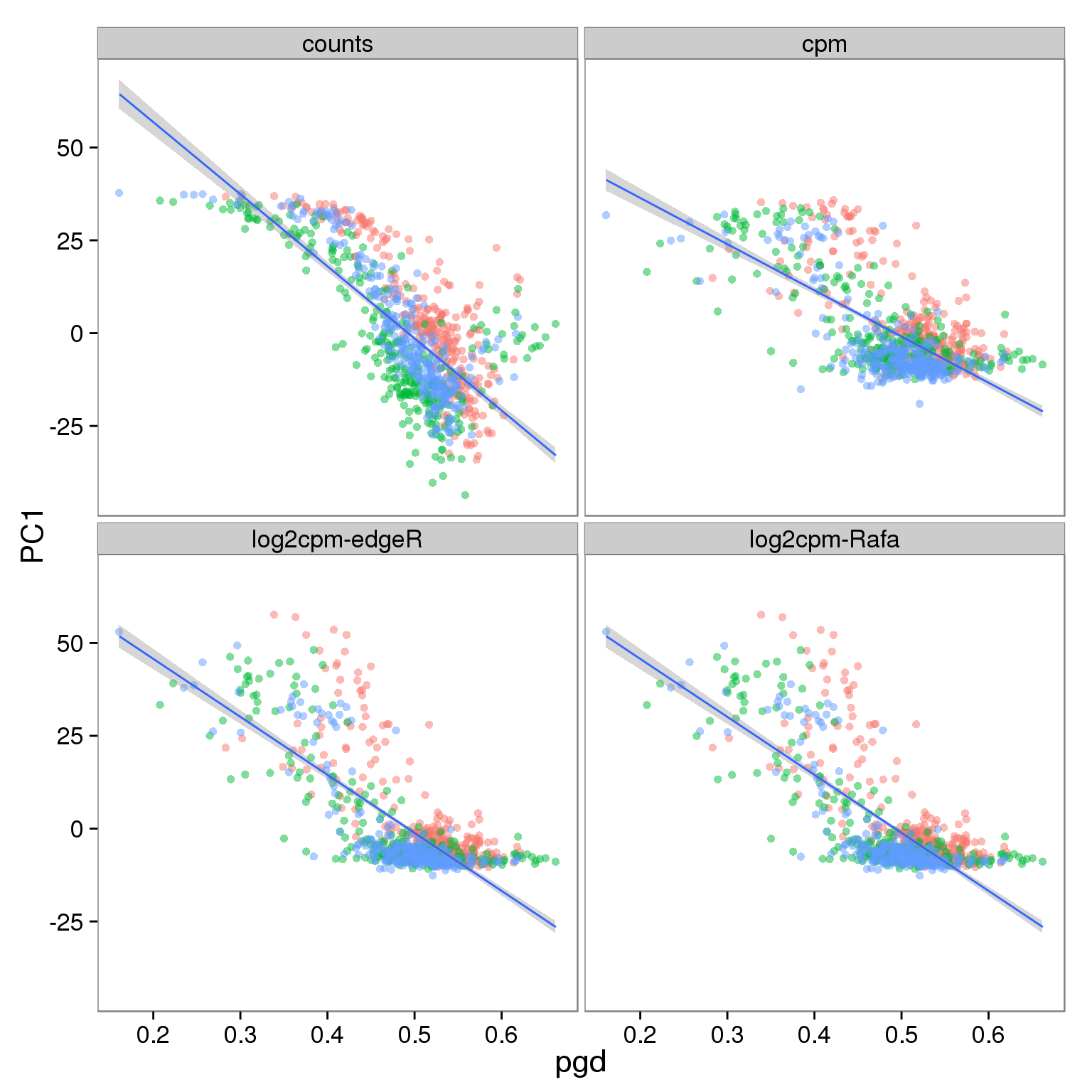
Effect of processing on molecules using ubiquitous genes:
p_molecules_filter <- p_reads_all %+% d_full[d_full$type == "molecules" & d_full$genes == "ubiq", ]
p_molecules_filter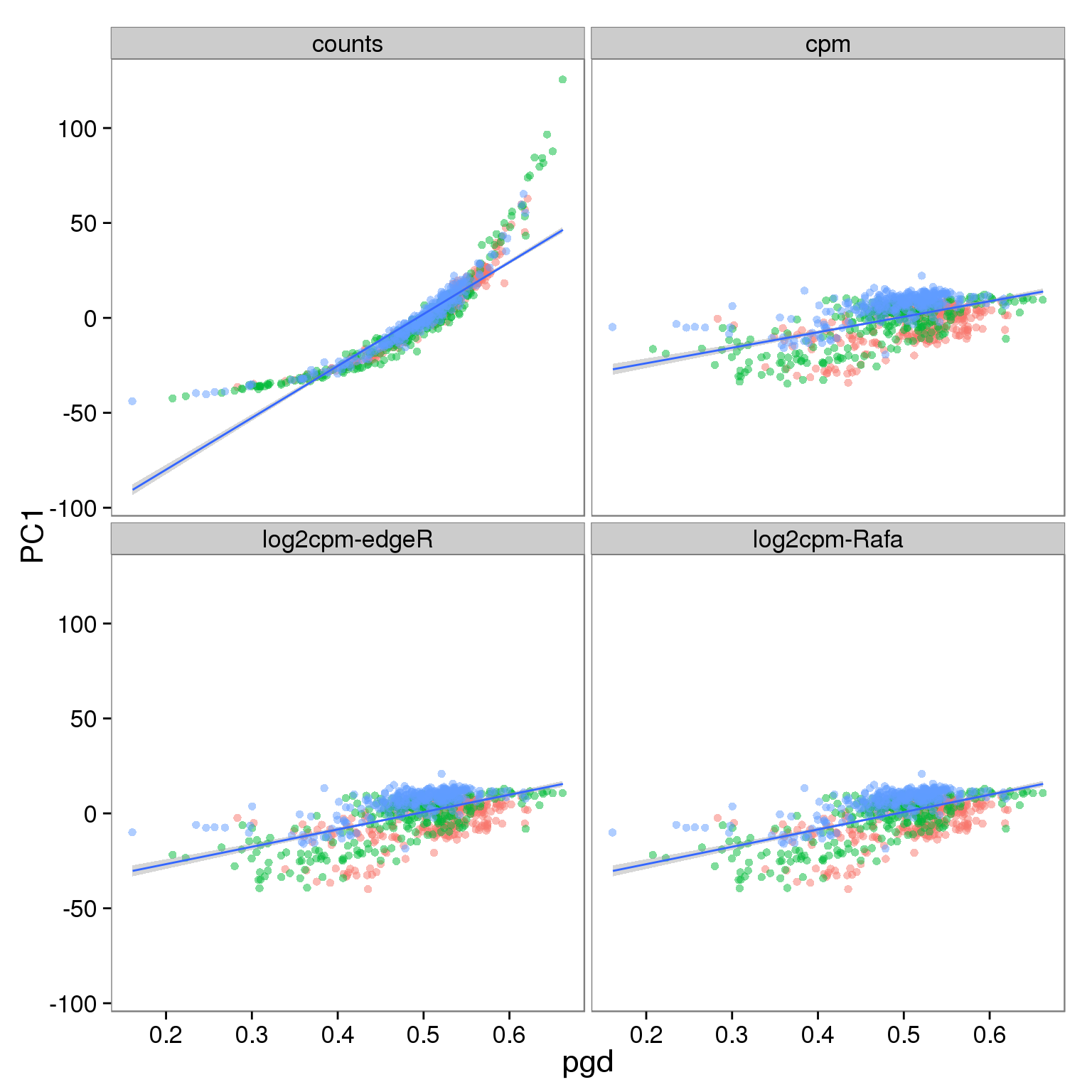
Conclusions
- PC1 is largely affected by cell quality, especially when PCA is performed using only ubiquitously expressed genes
- The correlation is stronger in reads data versus molecules
Session information
sessionInfo()R version 3.2.0 (2015-04-16)
Platform: x86_64-unknown-linux-gnu (64-bit)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] testit_0.4 cowplot_0.3.1 ggplot2_1.0.1 tidyr_0.2.0 edgeR_3.10.2
[6] limma_3.24.9 knitr_1.10.5
loaded via a namespace (and not attached):
[1] Rcpp_0.12.0 magrittr_1.5 MASS_7.3-40 munsell_0.4.2
[5] colorspace_1.2-6 stringr_1.0.0 httr_0.6.1 plyr_1.8.3
[9] tools_3.2.0 grid_3.2.0 gtable_0.1.2 htmltools_0.2.6
[13] yaml_2.1.13 digest_0.6.8 reshape2_1.4.1 formatR_1.2
[17] codetools_0.2-11 bitops_1.0-6 RCurl_1.95-4.6 evaluate_0.7
[21] rmarkdown_0.6.1 labeling_0.3 stringi_0.4-1 scales_0.2.4
[25] proto_0.3-10