Mixed effect model for batch correction - limma
Joyce Hsiao
2015-11-11
Last updated: 2015-12-11
Code version: 7a25b2aef5c8329d4b5394cce595bbfc2b5b0c3e
Objective
Compare per gene batch effect correction versus genewide batch effect correct using the [filtered data].
Setup
source("functions.R")
library("limma")
library("edgeR")Warning: package 'edgeR' was built under R version 3.2.2library(ggplot2)
theme_set(theme_bw(base_size = 16))Data preparation
Input quality single cells
quality_single_cells <- read.table("../data/quality-single-cells.txt",
header = TRUE,
stringsAsFactors = FALSE)
str(quality_single_cells)'data.frame': 567 obs. of 1 variable:
$ NA19098.r1.A01: chr "NA19098.r1.A02" "NA19098.r1.A04" "NA19098.r1.A05" "NA19098.r1.A06" ...Input annotation
anno <- read.table("../data/annotation.txt",
header = TRUE,
stringsAsFactors = FALSE)
anno_filter <- anno[ which(anno$sample_id %in% quality_single_cells[[1]]), ]
dim(anno_filter)[1] 567 5*Molecule counts
Input molecule counts.
molecules <- read.table("../data/molecules.txt",
header = TRUE,
stringsAsFactors = FALSE)
dim(molecules)[1] 18568 864Filter cells.
molecules_filter <- molecules[ , which(colnames(molecules) %in% quality_single_cells[[1]])]
dim(molecules_filter)[1] 18568 567Filter genes.
molecules_cpm_mean <- rowMeans(cpm(molecules_filter, log = TRUE))
lower_exp_cutoff <- 2
genes_pass_filter <- rownames(molecules_filter)[molecules_cpm_mean > lower_exp_cutoff]overexpressed_rows <- apply(molecules_filter, 1, function(x) any(x >= 1024))
overexpressed_genes <- rownames(molecules_filter)[overexpressed_rows]
genes_pass_filter <- setdiff(genes_pass_filter, overexpressed_genes)molecules_filter <- molecules_filter[rownames(molecules_filter) %in% genes_pass_filter, ]*Correct for collision probability
molecules_collision <- -1024 * log(1 - molecules_filter / 1024)*Molecules single cell endogeneous
ercc_rows <- grepl("ERCC", rownames(molecules_filter))
molecules_cpm <- cpm(molecules_collision[!ercc_rows, ], log = TRUE)*Linear transformation
molecules_cpm_ercc <- cpm(molecules_collision[ercc_rows, ], log = TRUE)
ercc <- read.table("../data/expected-ercc-molecules.txt", header = TRUE,
stringsAsFactors = FALSE)
ercc <- ercc[ercc$id %in% rownames(molecules_cpm_ercc), ]
ercc <- ercc[order(ercc$id), ]
ercc$log2_cpm <- cpm(ercc$ercc_molecules_well, log = TRUE)molecules_cpm_trans <- molecules_cpm
molecules_cpm_trans[, ] <- NA
intercept <- numeric(length = ncol(molecules_cpm_trans))
slope <- numeric(length = ncol(molecules_cpm_trans))
for (i in 1:ncol(molecules_cpm_trans)) {
fit <- lm(molecules_cpm_ercc[, i] ~ ercc$log2_cpm)
intercept[i] <- fit$coefficients[1]
slope[i] <- fit$coefficients[2]
# Y = mX + b -> X = (Y - b) / m
molecules_cpm_trans[, i] <- (molecules_cpm[, i] - intercept[i]) / slope[i]
}
dim(molecules_cpm_trans)[1] 10564 567Remove unwanted variation
Load the Humanzee package
if (!require(Humanzee, quietly = TRUE)) {
library(devtools)
install_github("jhsiao999/Humanzee")
library(Humanzee)
}Create design matrix and compute a consensus correlation coefficient using limma’s duplicateCorrelation function.
block <- anno_filter$batch
design <- model.matrix(~ 1 + individual, data = anno_filter)Compute correlation between replicates.
dup_corrs_file <- "../data/dup-corrs.rda"
if (file.exists(dup_corrs_file)) {
load(dup_corrs_file)
} else{
dup_corrs <- duplicateCorrelation(molecules_cpm_trans,
design = design, block = block)
save(dup_corrs, file = dup_corrs_file)
}
str(dup_corrs)List of 3
$ consensus.correlation: num 0.0367
$ cor : num 0.0367
$ atanh.correlations : num [1:10564] 0.00133 0.12879 0.10788 -0.00138 0.01398 ...Restrict correlation to between -1 and 1.
corrs_vec <- pmin(dup_corrs$atanh.correlations, 1)Distribution of per-gene cell-to-cell correlation across batches.
ggplot(data.frame(corrs = corrs_vec ),
aes(x = corrs) ) +
geom_histogram() +
geom_vline(data = data.frame(corr = dup_corrs$cor),
aes(xintercept = corr,
colour = "red"),
show_guide = FALSE)stat_bin: binwidth defaulted to range/30. Use 'binwidth = x' to adjust this.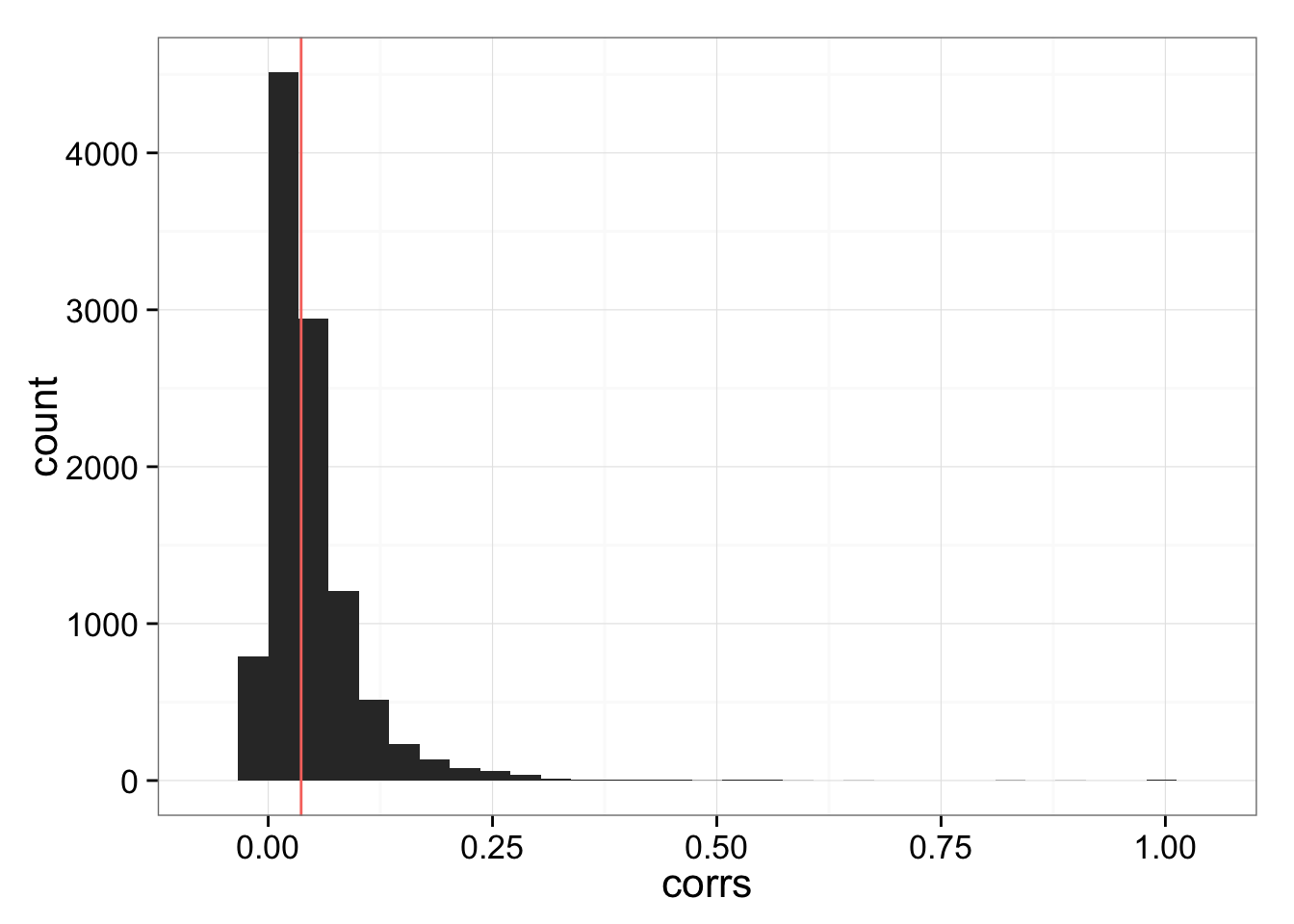
Gene-wise correction
if (file.exists("../data/limma-crossed-per-gene.rda")) {
load("../data/limma-crossed-per-gene.rda")
} else {
gls_fit_per_gene <- Humanzee::ruv_mixed_model(molecules_cpm_trans,
ndups = 1,
per_gene = FALSE,
design = design, block = block,
correlation = corrs_vec)
save(gls_fit_per_gene, file = "../data/limma-crossed-per-gene.rda")
}Compute expression levels after removing variation due to random effects.
molecules_final_per_gene <- t( design %*% t(gls_fit_per_gene$coef) ) + gls_fit_per_gene$residpca_final_per_gene <- run_pca(molecules_final_per_gene)
pca_final_plot_per_gene <- plot_pca(pca_final_per_gene$PCs, explained = pca_final_per_gene$explained,
metadata = anno_filter, color = "individual",
shape = "replicate") +
labs(title = "Molecules (batch corrected) for single cells")
pca_final_plot_per_gene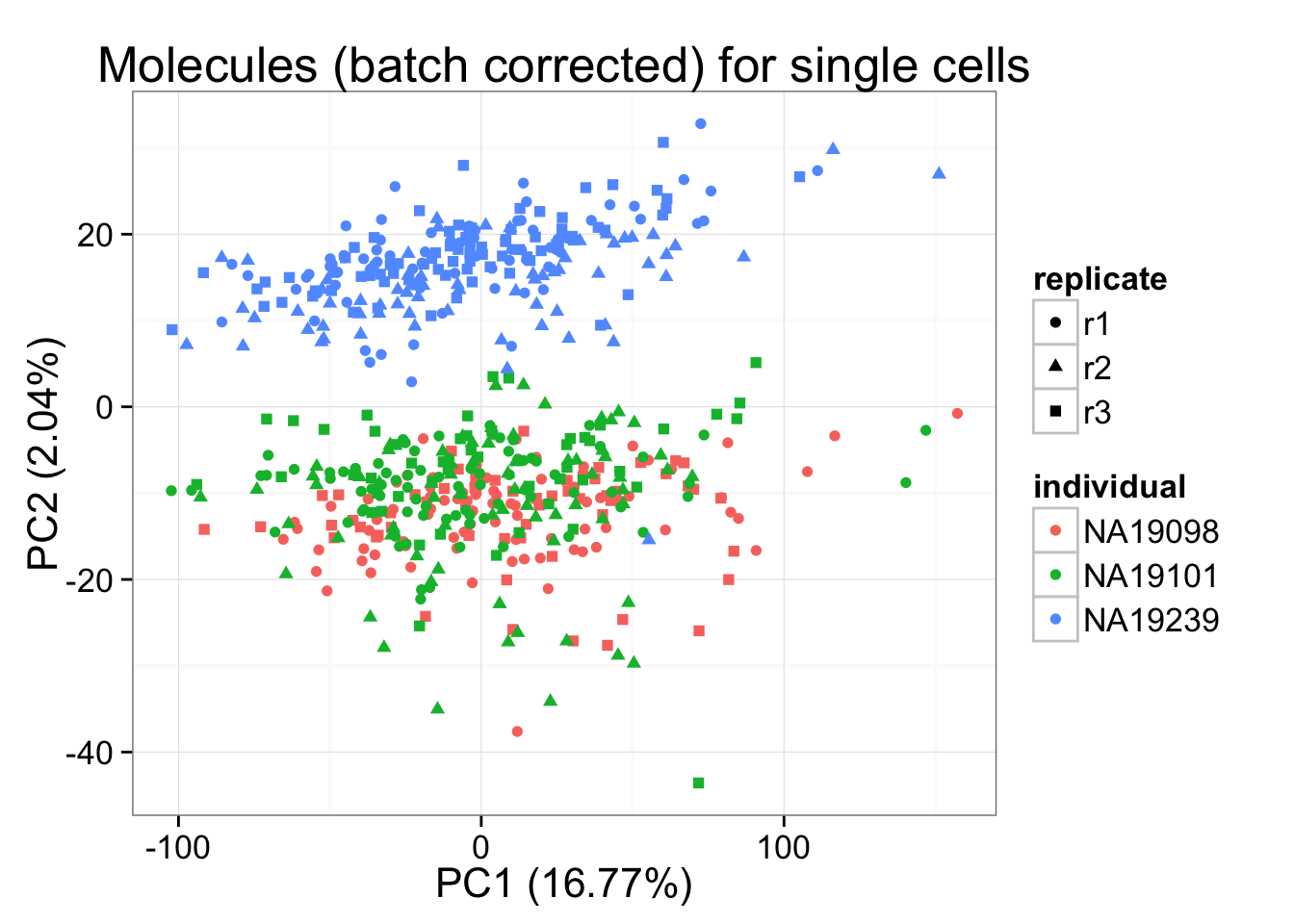
Gene-wide correction
if (file.exists("../data/limma-crossed.rda")) {
load("../data/limma-crossed.rda")
} else {
gls_fit <- Humanzee::ruv_mixed_model(molecules_cpm_trans,
ndups = 1,
per_gene = FALSE,
design = design, block = block,
correlation = dup_corrs$cor)
save(gls_fit, file = "../data/limma-crossed.rda")
}Compute expression levels after removing variation due to random effects.
molecules_final <- t( design %*% t(gls_fit$coef) ) + gls_fit$residpca_final <- run_pca(molecules_final)
pca_final_plot <- plot_pca(pca_final$PCs, explained = pca_final$explained,
metadata = anno_filter, color = "individual",
shape = "replicate") +
labs(title = "Molecules (batch corrected) for single cells")
pca_final_plot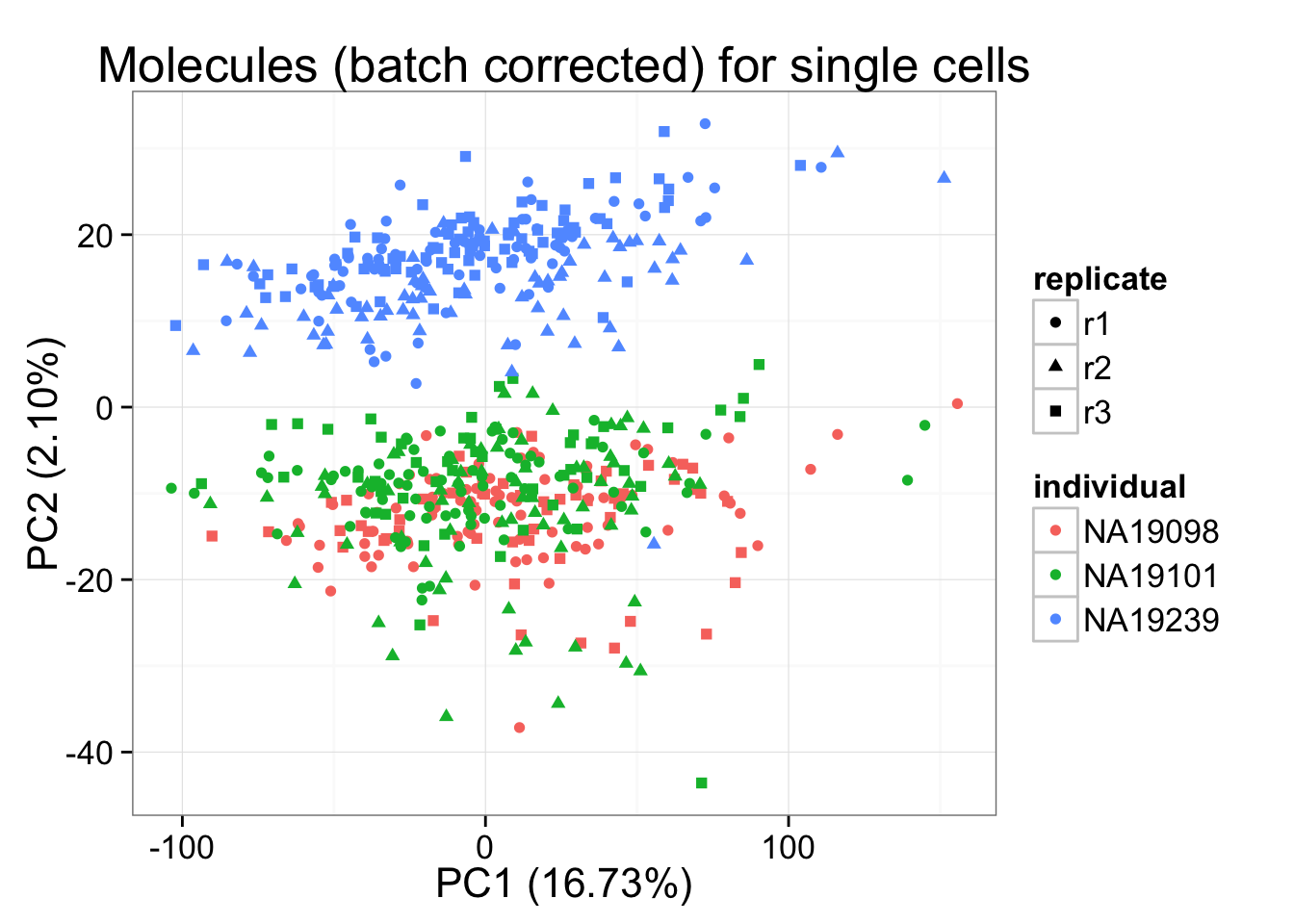
Expression ranking
*Gene ranks per cell are consistent between the gene-wise correction and the gene-wide correction methods. There seems to be more fluctuations in the correlation between the two methods in cell ranks per gene, which suggests a possible impact on the analysis comparing coefficients of variations between genes.
Per-cell correlation in expression levels
cell_corr <- sapply(1:NCOL(molecules_final), function(ii_cell) {
cor(molecules_final[ , ii_cell],
molecules_final_per_gene[ , ii_cell], method = "spearman")
})
summary(cell_corr) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.9665 0.9958 0.9968 0.9964 0.9977 0.9992 Per-gene correlation in expression levels
gene_corr <- sapply(1:NROW(molecules_final), function(ii_gene) {
cor(molecules_final[ii_gene, ],
molecules_final_per_gene[ii_gene, ], method = "spearman")
})
summary(gene_corr) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.6550 0.9972 0.9990 0.9974 0.9998 1.0000 Per-gene correlation in expression levels for each individual
gene_corr_individual <- lapply(1:3, function(ii_individual) {
which_individual <- anno_filter$individual == unique(anno_filter$individual)[ii_individual]
df <- molecules_final[ , which_individual]
df_per_gene <- molecules_final_per_gene[ , which_individual]
cell_corr <- sapply(1:NROW(molecules_final), function(ii_gene) {
cor(df[ ii_gene, ],
df_per_gene[ ii_gene, ], method = "spearman")
})
cell_corr
})
lapply(gene_corr_individual, summary)[[1]]
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.7182 0.9974 0.9991 0.9973 0.9997 1.0000
[[2]]
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.8431 0.9969 0.9989 0.9971 0.9997 1.0000
[[3]]
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.5158 0.9972 0.9990 0.9971 0.9997 1.0000 Session information
sessionInfo()R version 3.2.1 (2015-06-18)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X 10.10.5 (Yosemite)
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] testit_0.4 Humanzee_0.1.0 ggplot2_1.0.1 edgeR_3.10.5
[5] limma_3.24.15 knitr_1.11
loaded via a namespace (and not attached):
[1] Rcpp_0.12.2 magrittr_1.5 MASS_7.3-45 munsell_0.4.2
[5] colorspace_1.2-6 stringr_1.0.0 plyr_1.8.3 tools_3.2.1
[9] grid_3.2.1 gtable_0.1.2 htmltools_0.2.6 yaml_2.1.13
[13] digest_0.6.8 reshape2_1.4.1 formatR_1.2.1 evaluate_0.8
[17] rmarkdown_0.8.1 labeling_0.3 stringi_1.0-1 scales_0.3.0
[21] proto_0.3-10