Identifying single cells
John Blischak
2017-08-23
Last updated: 2018-07-01
Code version: 5eab055
Link to Yoruba samples on Coriell
Setup
library("cowplot")
library("dplyr")
library("ggplot2")
library("stringr")
library("tidyr")
theme_set(theme_cowplot())
library("Biobase")Import data.
eset <- readRDS("../data/eset.rds")
(chips <- unique(eset$experiment)) [1] "02192018" "02202018" "02212018" "02222018" "02242018" "02262018"
[7] "02272018" "02282018" "03012018" "03052018" "03062018" "03072018"
[13] "03162017" "03172017" "03232017" "03302017" "03312017" "04052017"
[19] "04072017" "04132017" "04142017" "04202017" "08102017" "08112017"
[25] "08142017" "08152017" "08162017" "08182017" "08212017" "08222017"
[31] "08232017" "08242017" "08282017" "08292017" "08302017" "08312017"
[37] "09252017" "09262017" "09272017" "10022017" "10042017" "10052017"
[43] "10062017" "10092017" "10102017" "10112017" "10122017" "10132017"
[49] "10162017" "10172017" "10302017" "11022017" "11032017" "11062017"
[55] "11072017" "11082017" "11092017" "11102017" "11132017" "11142017"
[61] "11152017" "11162017" "11172017" "11202017" "11212017" "11272017"
[67] "11282017" "11292017" "11302017" "12012017" "12042017" "12052017"
[73] "12062017" "12072017" "12082017" "12112017" "12122017" "12132017"
[79] "12142017"esetExpressionSet (storageMode: lockedEnvironment)
assayData: 54616 features, 7584 samples
element names: exprs
protocolData: none
phenoData
sampleNames: 02192018-A01 02192018-A02 ... 12142017-H12 (7584
total)
varLabels: experiment well ... valid_id (40 total)
varMetadata: labelDescription
featureData
featureNames: ENSG00000000003 ENSG00000000005 ... WBGene00235374
(54616 total)
fvarLabels: chr start ... source (6 total)
fvarMetadata: labelDescription
experimentData: use 'experimentData(object)'
Annotation: How many of the identities are plausible?
table(eset$valid_id)
FALSE TRUE
124 7460 mean(eset$valid_id)[1] 0.9836498Plot per C1 chip
anno <- pData(eset)
df_e <- anno %>%
group_by(experiment) %>%
summarize(i1 = unique(individual.1),
i2 = unique(individual.2),
i3 = unique(individual.3),
i4 = unique(individual.4))
op <- par(cex = 2, las = 2, mfrow = c(ceiling(length(chips) / 5), 5))
for (i in 1:nrow(df_e)) {
e <- df_e$experiment[i]
n1 <- sum(anno$chip_id[anno$experiment == e] == df_e$i1[i])
n2 <- sum(anno$chip_id[anno$experiment == e] == df_e$i2[i])
n3 <- sum(anno$chip_id[anno$experiment == e] == df_e$i3[i])
n4 <- sum(anno$chip_id[anno$experiment == e] == df_e$i4[i])
n_other <- 96 - sum(anno$chip_id[anno$experiment == e] %in%
as.character(df_e[i, ]))
barplot(c(n1, n2, n3, n4, n_other), main = sprintf("C1 chip %s", e),
names.arg = c(df_e$i1[i], df_e$i2[i], df_e$i3[i],df_e$i4[i], "Other"),
ylab = "Number of single cells")
}
par(op)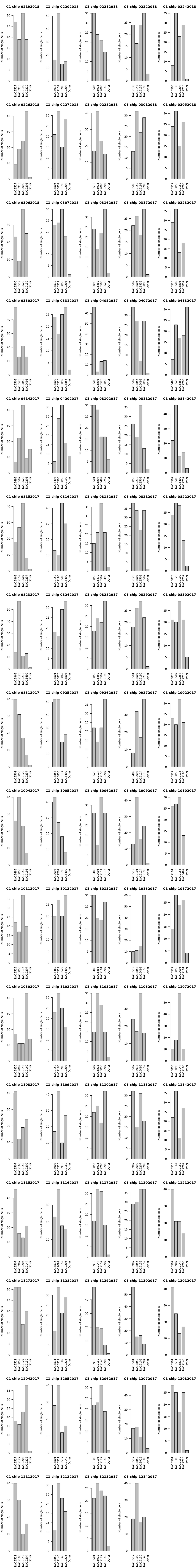
Colored by individual.
anno$valid_id_display <- factor(anno$valid_id, levels = c(TRUE, FALSE),
labels = c("Valid", "Invalid"))
ggplot(anno, aes(x = valid_id_display, fill = chip_id)) +
geom_bar() +
facet_wrap(~experiment) +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1),
legend.position = "bottom") +
labs(x = "", y = "Number of single cells",
title = "Valid vs. invalid assignments colored by individual")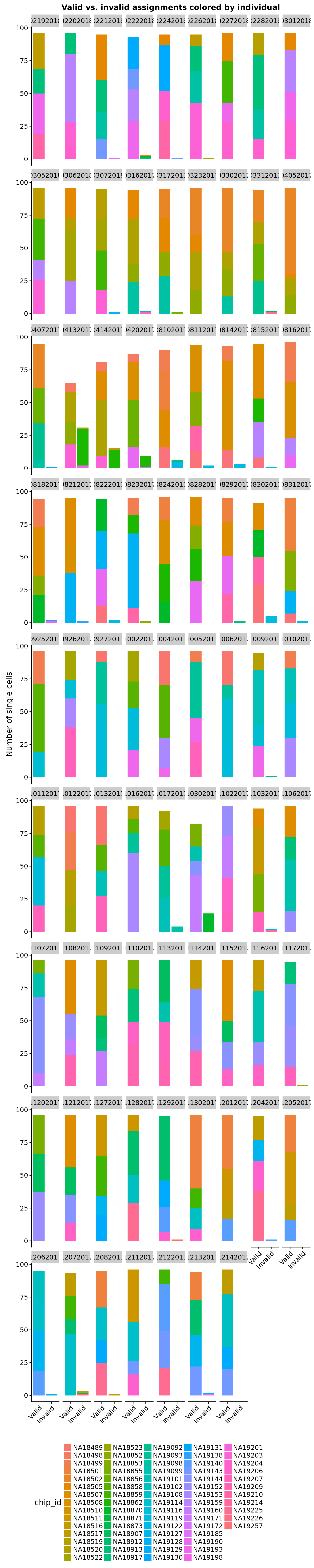
Expected versus observed cells
Each individual is expected to get 25% of the wells (i.e. 24) for each C1 chip it is on.
expected <- df_e %>% select(starts_with("i")) %>% unlist %>% table * 96 / 4
expected <- as.data.frame(expected)
colnames(expected) <- c("ind", "count")
observed <- table(anno$chip_id)
observed <- as.data.frame(observed)
colnames(observed) <- c("ind", "count")
df_obs <- merge(expected, observed, by = "ind", suffixes = c(".exp", ".obs"),
all.x = TRUE, sort = TRUE)
# Some individuals were assigned zero cells
df_obs$count.obs[is.na(df_obs$count.obs)] <- 0
df_obs ind count.exp count.obs
1 NA18489 120 110
2 NA18498 72 21
3 NA18499 216 166
4 NA18501 216 280
5 NA18502 144 231
6 NA18505 216 164
7 NA18507 360 420
8 NA18508 144 210
9 NA18511 120 149
10 NA18516 96 123
11 NA18517 192 139
12 NA18519 168 121
13 NA18520 168 172
14 NA18522 144 144
15 NA18852 144 103
16 NA18853 96 90
17 NA18855 120 108
18 NA18856 72 92
19 NA18858 192 188
20 NA18859 168 168
21 NA18862 96 135
22 NA18870 96 96
23 NA18907 120 115
24 NA18912 96 121
25 NA18913 216 191
26 NA19092 48 52
27 NA19093 120 124
28 NA19098 168 145
29 NA19099 120 122
30 NA19101 96 113
31 NA19102 96 99
32 NA19108 96 125
33 NA19114 120 100
34 NA19116 120 161
35 NA19127 96 85
36 NA19128 96 143
37 NA19130 144 134
38 NA19140 120 108
39 NA19143 144 112
40 NA19144 144 179
41 NA19152 192 155
42 NA19153 96 135
43 NA19159 168 188
44 NA19160 120 124
45 NA19190 120 116
46 NA19193 192 167
47 NA19203 168 156
48 NA19204 192 136
49 NA19206 120 85
50 NA19207 192 189
51 NA19209 96 102
52 NA19210 144 121
53 NA19225 96 113
54 NA19257 168 100# The max observed number of cells for any indvidual
max_cells <- max(df_obs$count.exp, df_obs$count.obs)
ggplot(df_obs, aes(x = count.exp, y = count.obs)) +
geom_text(aes(label = ind)) +
geom_abline(intercept = 0, slope = 1, color = "red", linetype = "dashed") +
xlim(0, max_cells) + ylim(0, max_cells) +
labs(x = "Expected number of cells per individual",
y = "Observed number of cells per individual",
title = "Observed versus expected number of cells")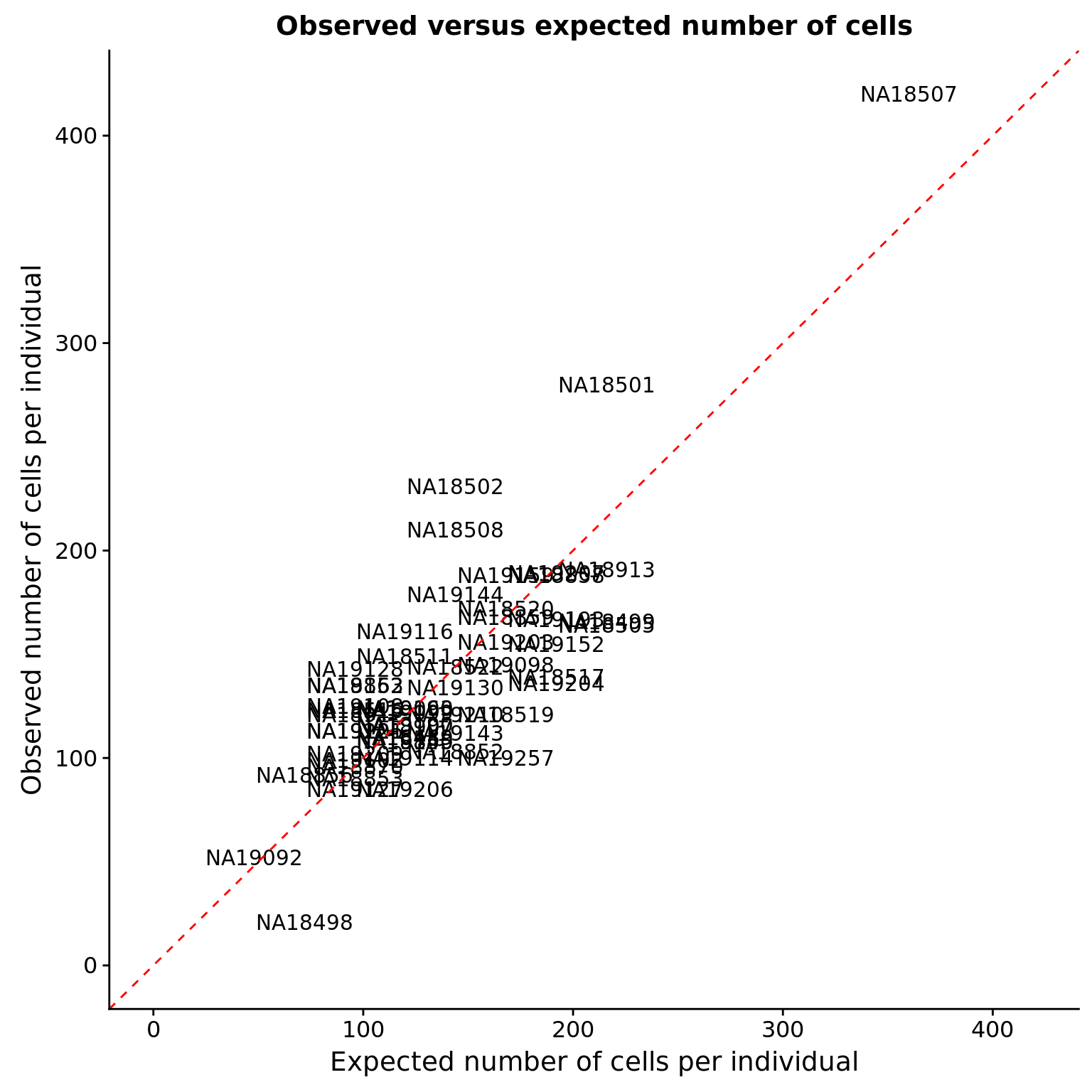
Unexpectedly high counts
observed %>% filter(count > 10, !(ind %in% expected$ind)) %>%
arrange(desc(count)) ind count
1 NA19119 17Top individuals per C1 chip
# Return all individuals with more than cutoff cells on the C1 chip
table_to_string <- function(x, cutoff = 5) {
tab <- sort(table(x), decreasing = TRUE)
tab <- tab[tab > cutoff]
string <- mapply(paste, names(tab), tab, MoreArgs = list(sep = " - "))
return(paste(string, collapse = "; "))
}
top <- anno %>%
group_by(experiment, individual.1, individual.2, individual.3, individual.4) %>%
summarize(Observed = table_to_string(chip_id))
knitr::kable(top, col.names = c("C1 chip", "Expected 1", "Expected 2",
"Expected 3", "Expected 4", "Observed (individual - number of cells)"))| C1 chip | Expected 1 | Expected 2 | Expected 3 | Expected 4 | Observed (individual - number of cells) |
|---|---|---|---|---|---|
| 02192018 | NA18517 | NA18913 | NA19193 | NA19210 | NA19193 - 31; NA18517 - 27; NA18913 - 19; NA19210 - 19 |
| 02202018 | NA18913 | NA19159 | NA19203 | NA19204 | NA19159 - 52; NA18913 - 16; NA19204 - 15; NA19203 - 13 |
| 02212018 | NA18505 | NA18913 | NA19098 | NA19143 | NA18505 - 35; NA18913 - 24; NA19098 - 21; NA19143 - 15 |
| 02222018 | NA19130 | NA19143 | NA19159 | NA19193 | NA19193 - 29; NA19130 - 24; NA19159 - 24; NA19143 - 16 |
| 02242018 | NA18505 | NA19130 | NA19204 | NA19210 | NA19130 - 35; NA19210 - 29; NA19204 - 23; NA18505 - 8 |
| 02262018 | NA18517 | NA18913 | NA19098 | NA19203 | NA19203 - 43; NA19098 - 24; NA18913 - 19; NA18517 - 9 |
| 02272018 | NA18505 | NA18859 | NA19193 | NA19204 | NA18859 - 32; NA19204 - 28; NA18505 - 21; NA19193 - 15 |
| 02282018 | NA18519 | NA18913 | NA19098 | NA19204 | NA18913 - 41; NA19098 - 23; NA18519 - 17; NA19204 - 15 |
| 03012018 | NA18505 | NA19159 | NA19193 | NA19203 | NA19159 - 32; NA19203 - 29; NA19193 - 22; NA18505 - 13 |
| 03052018 | NA18517 | NA18859 | NA19159 | NA19203 | NA18859 - 31; NA19203 - 26; NA18517 - 24; NA19159 - 15 |
| 03062018 | NA18505 | NA18519 | NA18522 | NA19159 | NA18522 - 39; NA19159 - 25; NA18505 - 23; NA18519 - 9 |
| 03072018 | NA18519 | NA18522 | NA18859 | NA19203 | NA18859 - 30; NA18522 - 24; NA18519 - 23; NA19203 - 18 |
| 03162017 | NA19098 | NA18852 | NA18505 | NA18520 | NA18520 - 34; NA19098 - 24; NA18505 - 22; NA18852 - 14 |
| 03172017 | NA18502 | NA18505 | NA18852 | NA19098 | NA19098 - 29; NA18505 - 26; NA18502 - 22; NA18852 - 18 |
| 03232017 | NA18520 | NA18502 | NA18505 | NA18852 | NA18502 - 36; NA18520 - 29; NA18852 - 18; NA18505 - 13 |
| 03302017 | NA18502 | NA18520 | NA18852 | NA19098 | NA18502 - 49; NA18852 - 21; NA18520 - 13; NA19098 - 13 |
| 03312017 | NA18502 | NA18520 | NA19092 | NA18856 | NA18856 - 28; NA19092 - 25; NA18502 - 24; NA18520 - 17 |
| 04052017 | NA18502 | NA18505 | NA18520 | NA18852 | NA18502 - 66; NA18852 - 14; NA18520 - 13 |
| 04072017 | NA18502 | NA18856 | NA19098 | NA19092 | NA18502 - 34; NA18856 - 27; NA19092 - 27; NA19098 - 7 |
| 04132017 | NA18498 | NA18520 | NA18852 | NA19203 | NA18862 - 28; NA18520 - 23; NA19203 - 18; NA18852 - 17; NA18498 - 7 |
| 04142017 | NA18498 | NA18507 | NA18520 | NA19203 | NA18520 - 43; NA18507 - 22; NA18862 - 14; NA19203 - 9; NA18498 - 7 |
| 04202017 | NA18498 | NA18508 | NA18856 | NA19190 | NA18856 - 36; NA18508 - 29; NA19190 - 16; NA18862 - 8; NA18498 - 6 |
| 08102017 | NA18501 | NA18507 | NA18499 | NA19257 | NA18501 - 30; NA18507 - 28; NA18499 - 16; NA19257 - 16 |
| 08112017 | NA18853 | NA19210 | NA18508 | NA19257 | NA18508 - 36; NA18853 - 26; NA19210 - 19; NA19257 - 13 |
| 08142017 | NA18507 | NA18508 | NA18499 | NA19257 | NA18508 - 46; NA18507 - 22; NA19257 - 14; NA18499 - 11 |
| 08152017 | NA18862 | NA19159 | NA18507 | NA19257 | NA18507 - 42; NA19159 - 27; NA18862 - 18; NA19257 - 8 |
| 08162017 | NA19159 | NA19190 | NA18508 | NA18499 | NA18508 - 43; NA18499 - 30; NA19159 - 13; NA19190 - 10 |
| 08182017 | NA18853 | NA18870 | NA18507 | NA18499 | NA18507 - 37; NA18499 - 21; NA18870 - 21; NA18853 - 15 |
| 08212017 | NA19128 | NA18507 | NA18508 | NA18507 | NA19128 - 38; NA18507 - 34; NA18508 - 23 |
| 08222017 | NA18870 | NA19128 | NA19190 | NA19257 | NA19128 - 29; NA19190 - 28; NA18870 - 24; NA19257 - 13 |
| 08232017 | NA18862 | NA19128 | NA19210 | NA18499 | NA19128 - 57; NA18862 - 14; NA18499 - 13; NA19210 - 11 |
| 08242017 | NA18501 | NA18870 | NA18862 | NA18508 | NA18508 - 33; NA18862 - 29; NA18501 - 18; NA18870 - 16 |
| 08282017 | NA18853 | NA18862 | NA18507 | NA19190 | NA19190 - 32; NA18862 - 24; NA18507 - 22; NA18853 - 18 |
| 08292017 | NA18501 | NA18507 | NA19190 | NA19210 | NA19190 - 29; NA18507 - 26; NA19210 - 22; NA18501 - 18 |
| 08302017 | NA18870 | NA18507 | NA19257 | NA19210 | NA19257 - 29; NA18870 - 21; NA19210 - 21; NA18507 - 20 |
| 08312017 | NA18501 | NA18853 | NA19128 | NA19257 | NA18501 - 40; NA18853 - 31; NA19128 - 17; NA19257 - 7 |
| 09252017 | NA18858 | NA18858 | NA19114 | NA18499 | NA18858 - 52; NA18499 - 25; NA19114 - 19 |
| 09262017 | NA18522 | NA19114 | NA19153 | NA19207 | NA19207 - 38; NA18522 - 22; NA19153 - 22; NA19114 - 14 |
| 09272017 | NA18489 | NA19093 | NA19114 | NA19116 | NA19116 - 39; NA19093 - 32; NA19114 - 17; NA18489 - 8 |
| 10022017 | NA18522 | NA18858 | NA19116 | NA19193 | NA19116 - 32; NA18522 - 23; NA19193 - 21; NA18858 - 20 |
| 10042017 | NA18489 | NA18858 | NA19153 | NA19193 | NA18858 - 40; NA18489 - 26; NA19153 - 23; NA19193 - 7 |
| 10052017 | NA19093 | NA19207 | NA19193 | NA18499 | NA19093 - 43; NA19207 - 27; NA19193 - 18; NA18499 - 8 |
| 10062017 | NA18489 | NA19093 | NA19114 | NA19116 | NA19114 - 34; NA18489 - 26; NA19116 - 26; NA19093 - 10 |
| 10092017 | NA18519 | NA19101 | NA19114 | NA19193 | NA19101 - 42; NA19193 - 24; NA19114 - 16; NA18519 - 13 |
| 10102017 | NA19101 | NA19116 | NA19153 | NA18499 | NA19153 - 30; NA19116 - 27; NA19101 - 26; NA18499 - 13 |
| 10112017 | NA18519 | NA18858 | NA19116 | NA19207 | NA19116 - 37; NA18519 - 22; NA19207 - 20; NA18858 - 17 |
| 10122017 | NA18489 | NA18519 | NA18522 | NA18499 | NA18499 - 29; NA18519 - 27; NA18489 - 20; NA18522 - 20 |
| 10132017 | NA18489 | NA18858 | NA19101 | NA19207 | NA18489 - 30; NA19207 - 27; NA18858 - 20; NA19101 - 19 |
| 10162017 | NA18519 | NA18858 | NA19093 | NA19153 | NA19153 - 60; NA19093 - 15; NA18858 - 11; NA18519 - 10 |
| 10172017 | NA18522 | NA18858 | NA19093 | NA19101 | NA18858 - 28; NA19101 - 26; NA19093 - 24; NA18522 - 14 |
| 10302017 | NA18855 | NA19099 | NA19144 | NA19160 | NA19160 - 43; NA18855 - 17; NA18870 - 14; NA19099 - 11; NA19144 - 11 |
| 11022017 | NA19152 | NA19160 | NA19206 | NA19207 | NA19160 - 32; NA19206 - 25; NA19152 - 23; NA19207 - 16 |
| 11032017 | NA18507 | NA18516 | NA18855 | NA19207 | NA18516 - 35; NA18855 - 29; NA18507 - 15; NA19207 - 15 |
| 11062017 | NA18507 | NA18913 | NA19099 | NA19152 | NA19099 - 39; NA18507 - 24; NA18913 - 17; NA19152 - 16 |
| 11072017 | NA18855 | NA19099 | NA19144 | NA19160 | NA19144 - 58; NA19099 - 18; NA18855 - 10; NA19160 - 10 |
| 11082017 | NA18507 | NA19160 | NA19152 | NA19209 | NA18507 - 41; NA19209 - 24; NA19152 - 19; NA19160 - 12 |
| 11092017 | NA18907 | NA18516 | NA18913 | NA19160 | NA18516 - 42; NA19160 - 27; NA18907 - 17; NA18913 - 10 |
| 11102017 | NA18855 | NA18913 | NA19206 | NA19209 | NA19209 - 32; NA18913 - 25; NA18855 - 22; NA19206 - 17 |
| 11132017 | NA18907 | NA19099 | NA19207 | NA19209 | NA18907 - 32; NA19207 - 31; NA19209 - 18; NA19099 - 15 |
| 11142017 | NA18516 | NA19144 | NA19152 | NA19209 | NA19144 - 36; NA19209 - 27; NA18516 - 22; NA19152 - 11 |
| 11152017 | NA18507 | NA18907 | NA19206 | NA19144 | NA18507 - 46; NA19144 - 21; NA18907 - 16; NA19206 - 13 |
| 11162017 | NA18516 | NA19099 | NA19152 | NA19206 | NA19099 - 39; NA18516 - 23; NA19152 - 18; NA19206 - 16 |
| 11172017 | NA18913 | NA19144 | NA19152 | NA19207 | NA19144 - 32; NA19152 - 31; NA18913 - 17; NA19207 - 15 |
| 11202017 | NA18907 | NA18855 | NA19152 | NA19152 | NA19152 - 37; NA18855 - 30; NA18907 - 29 |
| 11212017 | NA18507 | NA18907 | NA19144 | NA19206 | NA18507 - 40; NA18907 - 21; NA19144 - 21; NA19206 - 14 |
| 11272017 | NA18511 | NA18859 | NA19127 | NA19130 | NA18511 - 31; NA18859 - 31; NA19130 - 20; NA19127 - 14 |
| 11282017 | NA18511 | NA18912 | NA19102 | NA19225 | NA18912 - 34; NA19225 - 29; NA19102 - 21; NA18511 - 12 |
| 11292017 | NA18912 | NA19130 | NA19140 | NA19204 | NA18912 - 49; NA19130 - 20; NA19140 - 19; NA19204 - 7 |
| 11302017 | NA18501 | NA18859 | NA19102 | NA19204 | NA18501 - 56; NA19102 - 16; NA18859 - 15; NA19204 - 9 |
| 12012017 | NA18501 | NA18511 | NA18517 | NA19140 | NA18501 - 41; NA18511 - 25; NA19140 - 17; NA18517 - 13 |
| 12042017 | NA18517 | NA19127 | NA19204 | NA19225 | NA19225 - 38; NA19204 - 23; NA18517 - 18; NA19127 - 16 |
| 12052017 | NA18501 | NA18511 | NA18517 | NA19140 | NA18511 - 40; NA18501 - 28; NA19140 - 16; NA18517 - 12 |
| 12062017 | NA19102 | NA19108 | NA19127 | NA19140 | NA19127 - 31; NA19108 - 23; NA19102 - 22; NA19140 - 19 |
| 12072017 | NA18517 | NA18859 | NA18912 | NA19108 | NA19108 - 47; NA18859 - 18; NA18517 - 17; NA18912 - 11 |
| 12082017 | NA18501 | NA19108 | NA19130 | NA19225 | NA18501 - 28; NA19108 - 25; NA19225 - 25; NA19130 - 17 |
| 12112017 | NA18511 | NA19108 | NA19143 | NA19204 | NA18511 - 40; NA19108 - 30; NA19204 - 16; NA19143 - 10 |
| 12122017 | NA18859 | NA19140 | NA19143 | NA19225 | NA19140 - 36; NA19143 - 28; NA19225 - 21; NA18859 - 11 |
| 12132017 | NA18501 | NA18912 | NA19127 | NA19143 | NA18912 - 27; NA19127 - 24; NA19143 - 22; NA18501 - 21 |
| 12142017 | NA18517 | NA19102 | NA19130 | NA19143 | NA19102 - 40; NA19143 - 20; NA18517 - 19; NA19130 - 17 |
Potential reasons for failure
Sequencing depth
reason_raw <- ggplot(anno, aes(x = valid_id, y = raw)) +
geom_boxplot() +
scale_x_discrete(labels = c("Wrong", "Correct")) +
labs(x = "Predicted individual of origin",
y = "Raw number of reads",
title = "Raw number of reads")
reason_raw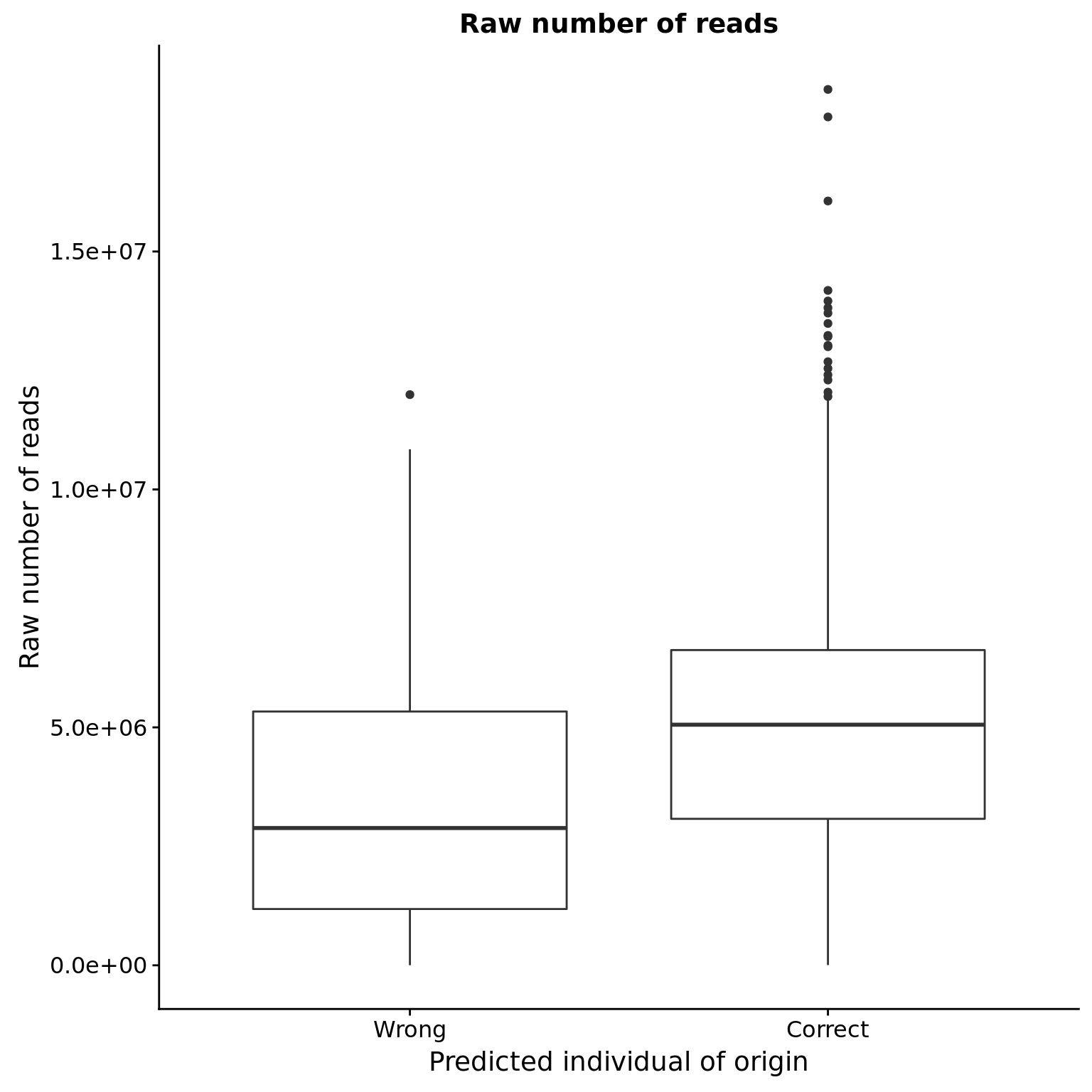
Percent mapped
reason_raw %+% aes(y = mapped / raw * 100) +
labs(y = "Percent of reads mapped",
title = "Percent of reads mapped")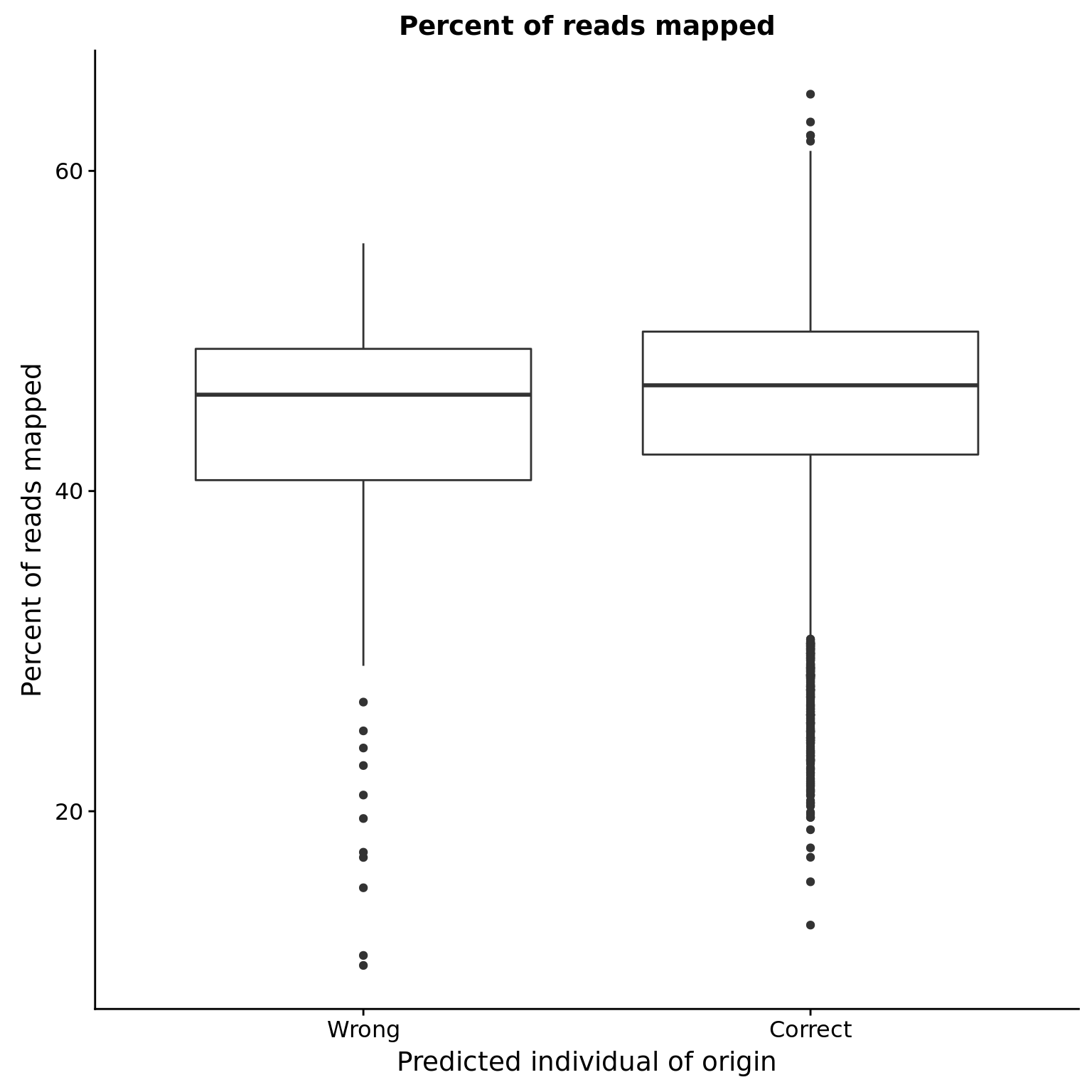
Cell number
barplot(with(anno, table(valid_id, cell_number)), beside = TRUE,
col = c("red", "black"),
xlab = "Number of cells in well", main = "Number of cells in well",
ylab = "Number of cells correct (black) versus wrong (red)")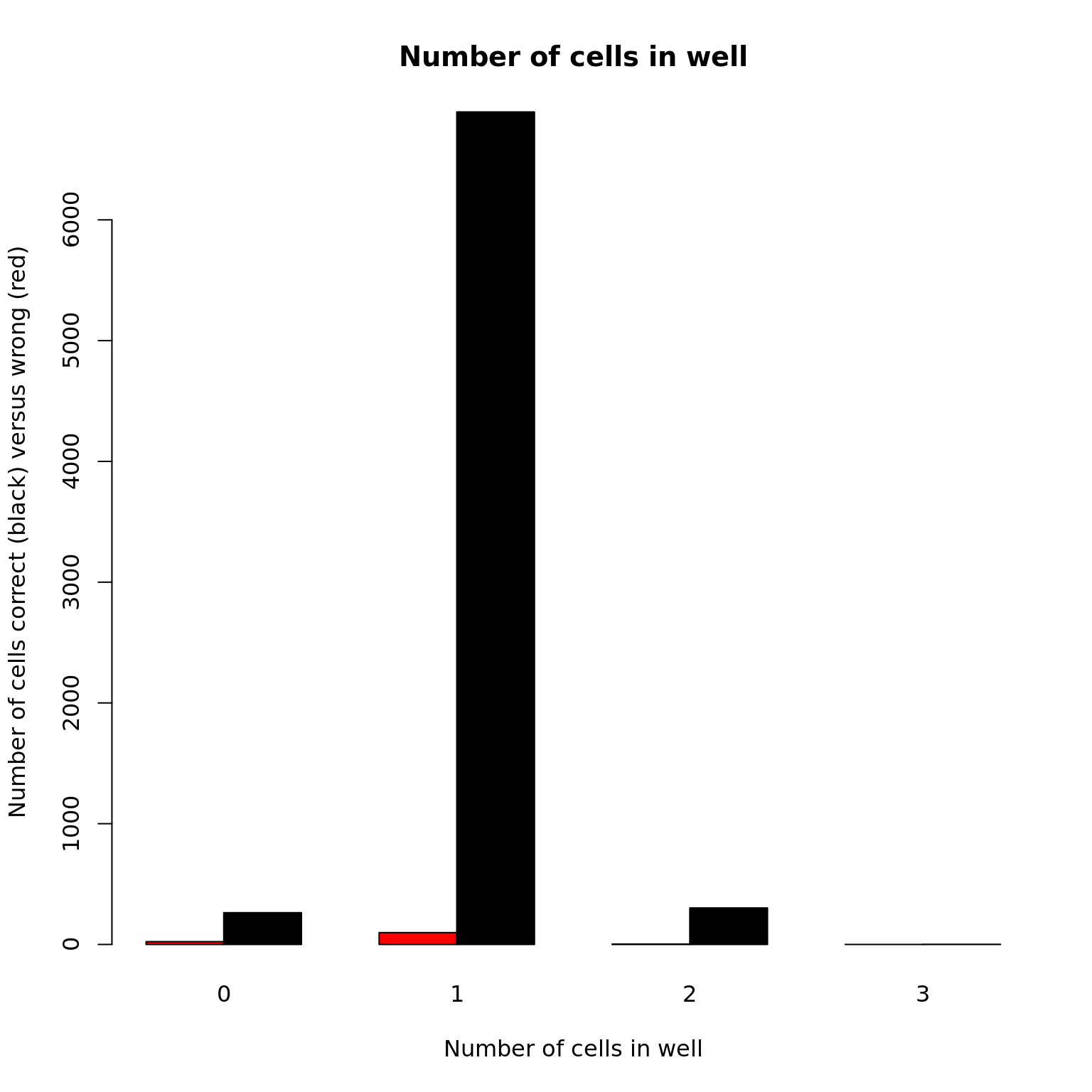
Concentration
reason_raw %+% aes(y = concentration) +
labs(y = "Concentration",
title = "Concentration")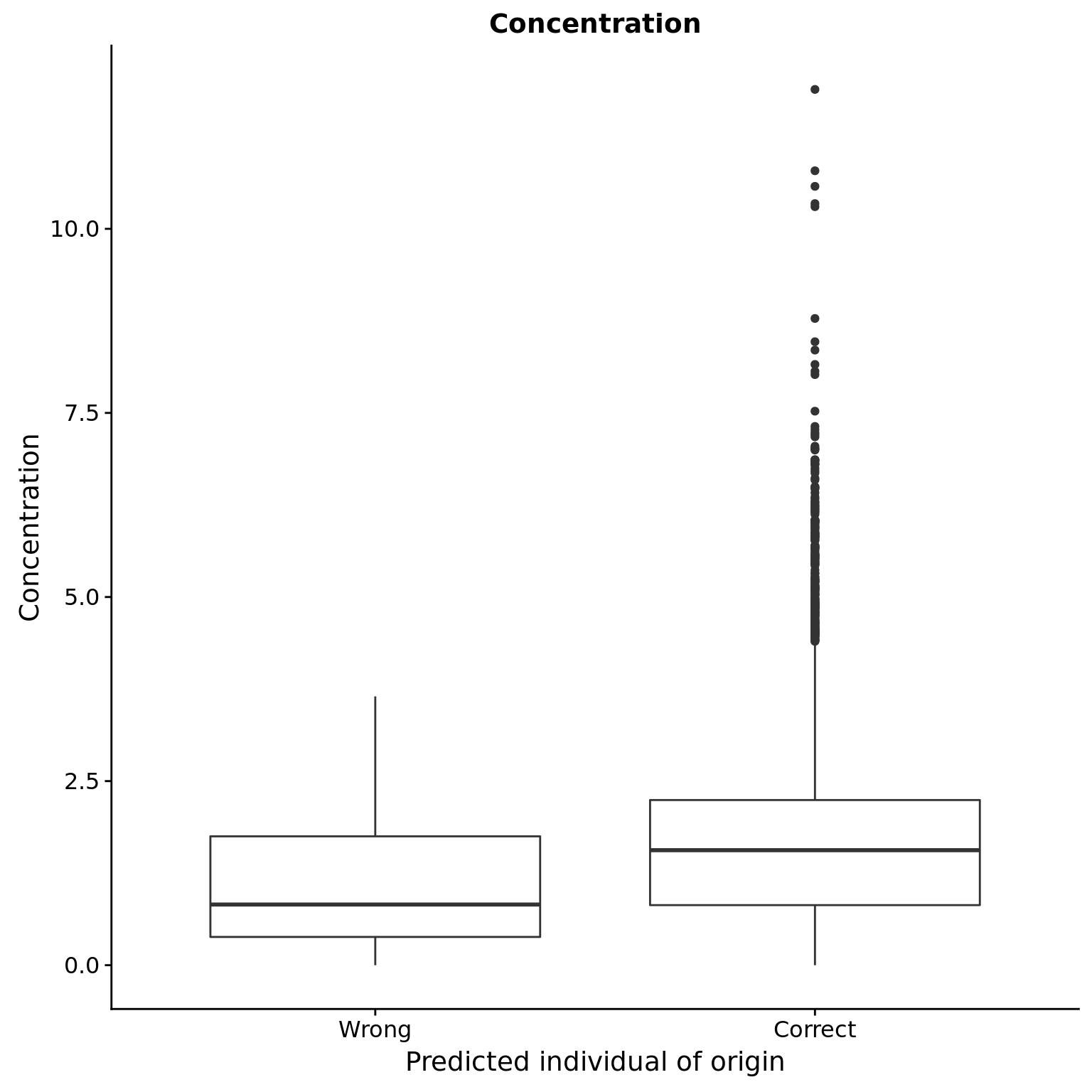
TRA-1-60
barplot(with(anno, table(valid_id, tra1.60)), beside = TRUE,
col = c("red", "black"),
xlab = "TRA-1-60", main = "TRA-1-60",
ylab = "Number of cells correct (black) versus wrong (red)")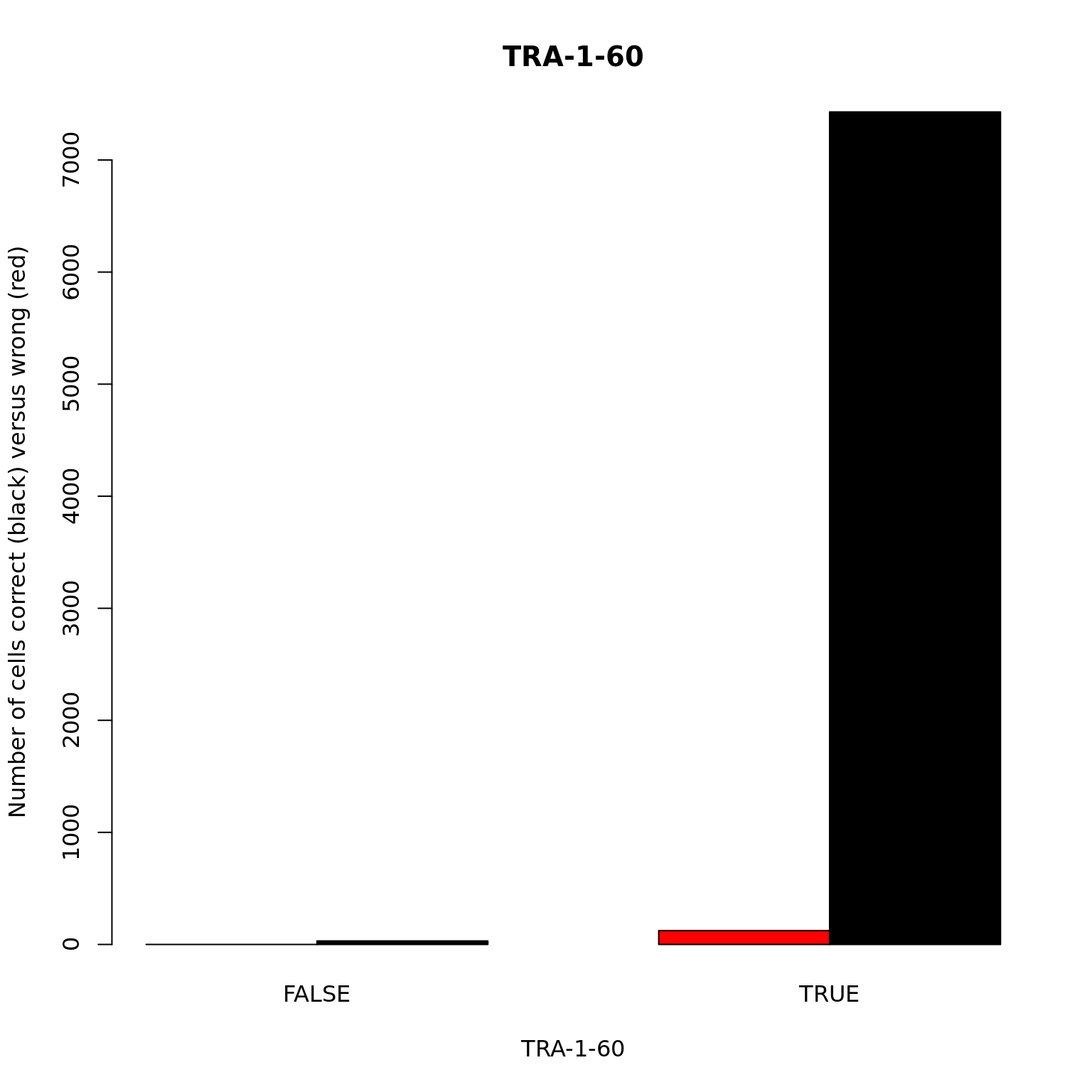
SNPs with at least one overlapping read
reason_raw %+% aes(y = snps_w_min) +
labs(y = "SNPs with at least one overlapping read",
title = "SNPs with at least one overlapping read")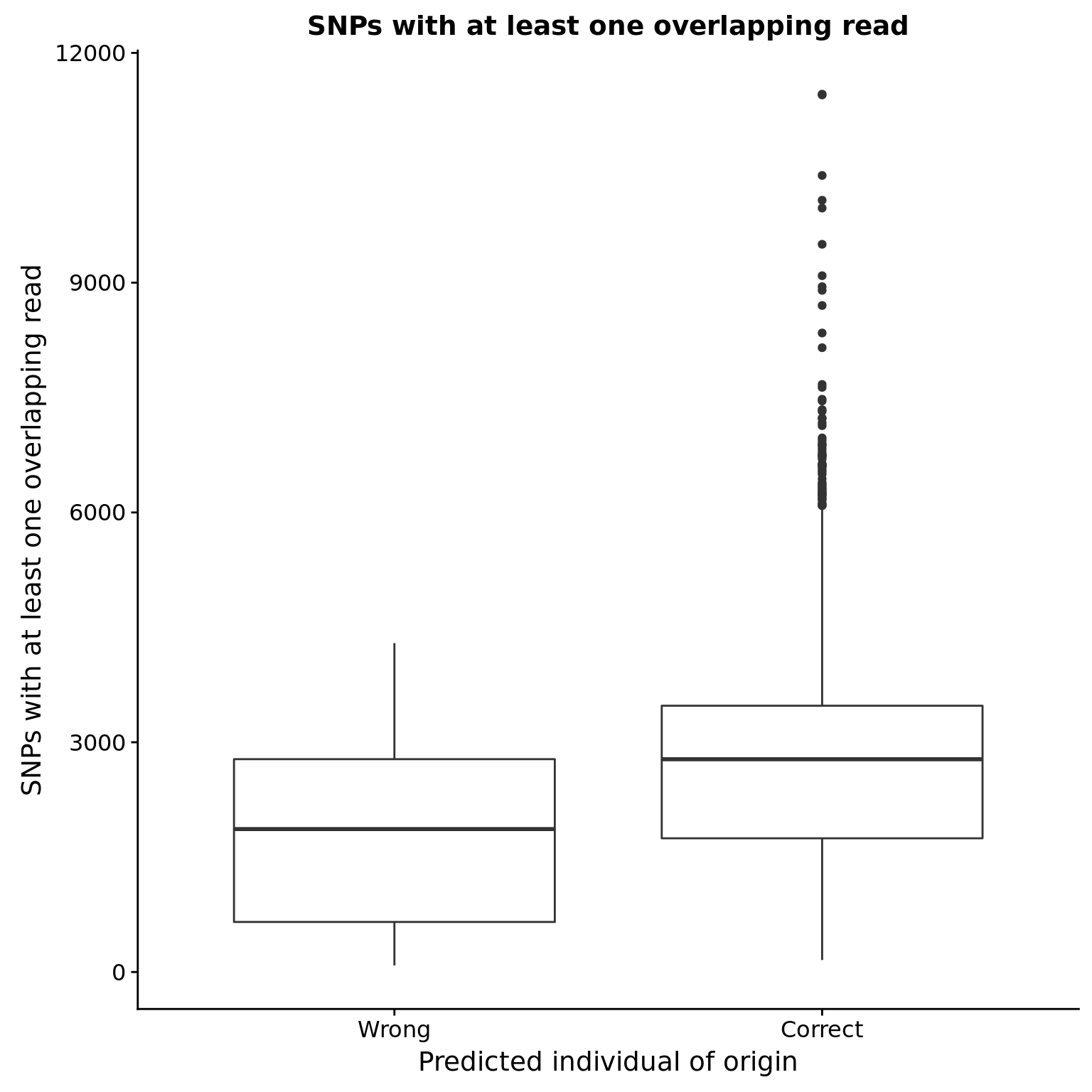
chipmix
reason_raw %+% aes(y = chipmix) +
labs(y = "chipmix", title = "chipmix")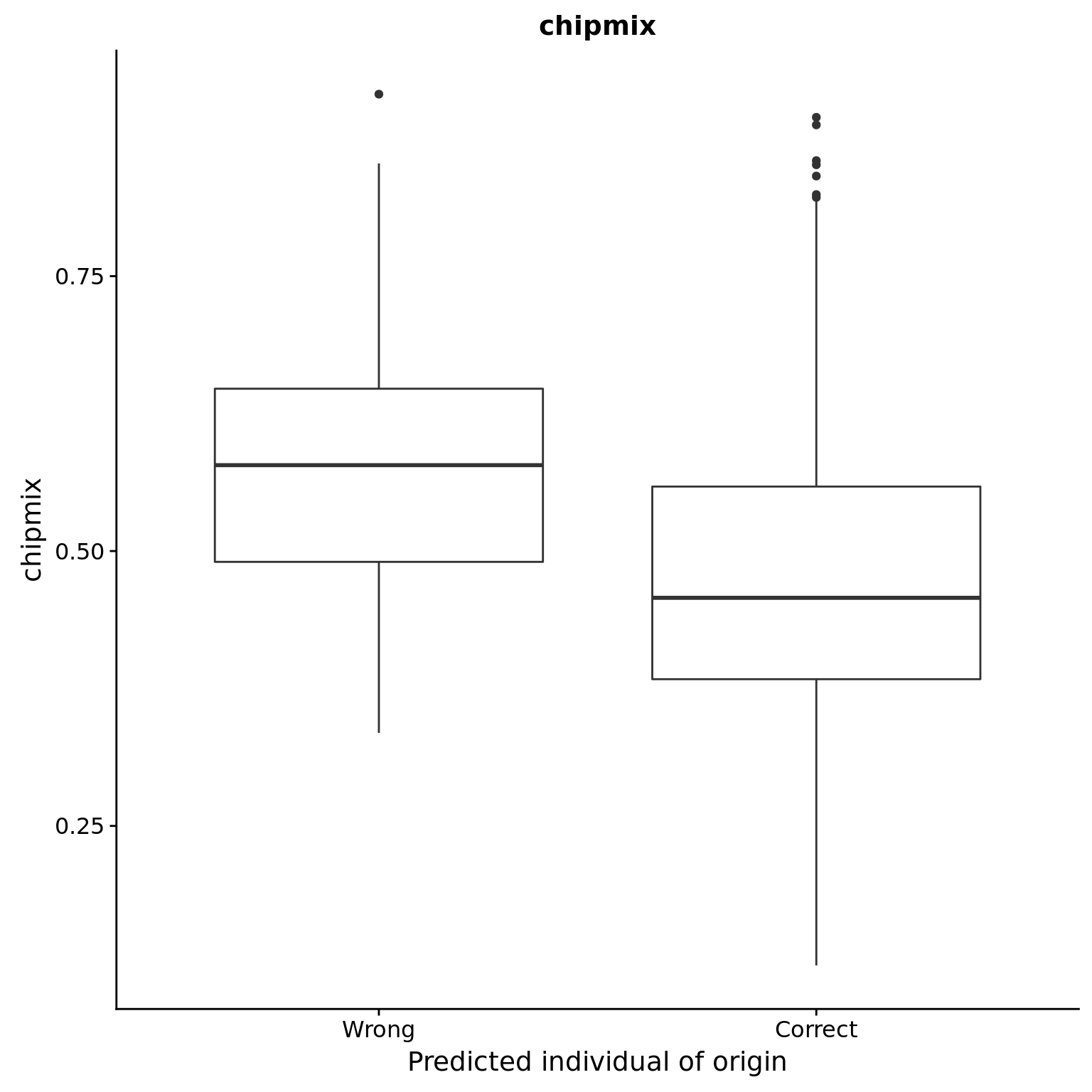
freemix
reason_raw %+% aes(y = freemix) +
labs(y = "freemix", title = "freemix")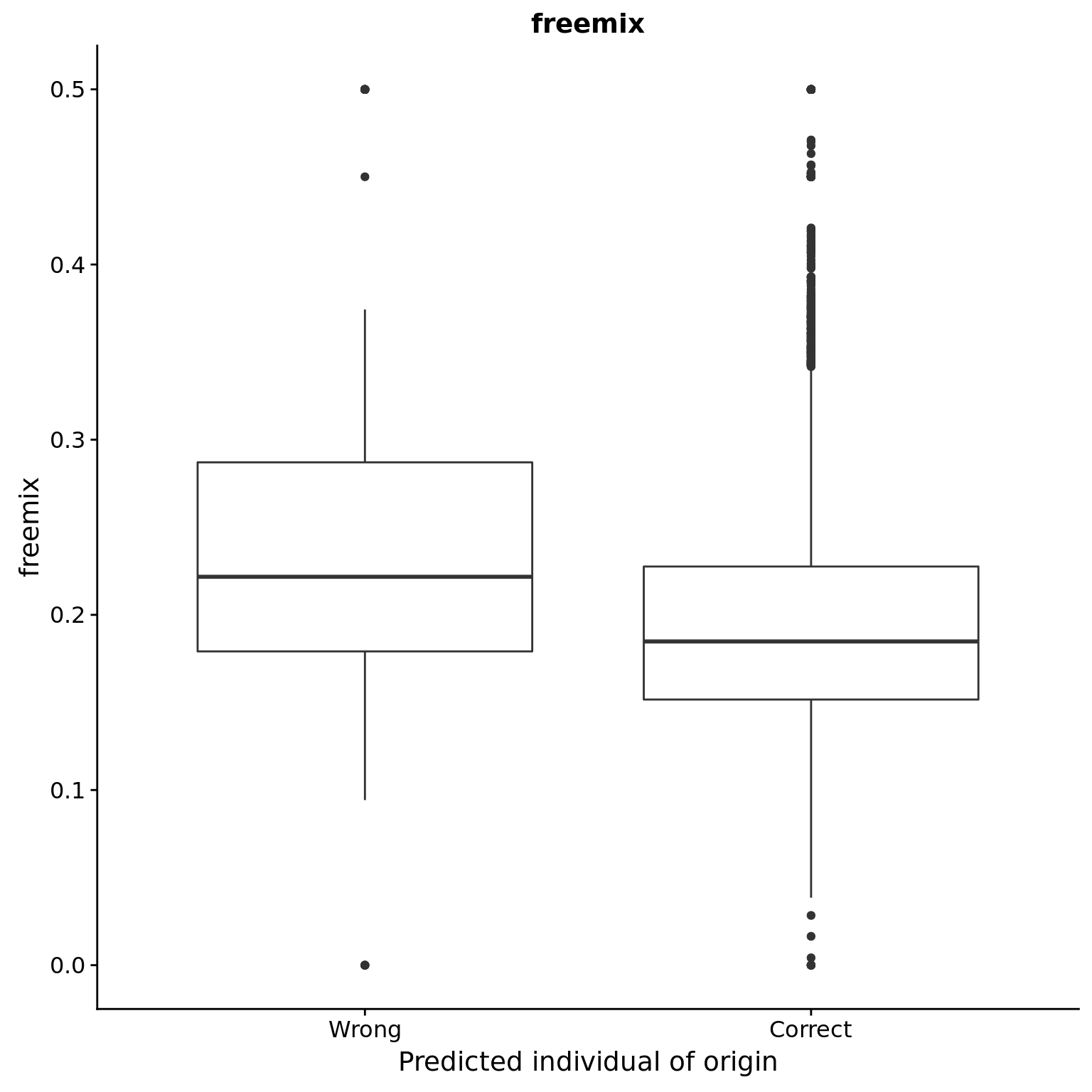
chipmix versus freemix
ggplot(anno, aes(x = freemix, y = chipmix, color = valid_id)) +
geom_point() +
facet_wrap(~valid_id) +
labs(title = "chipmix versus freemix")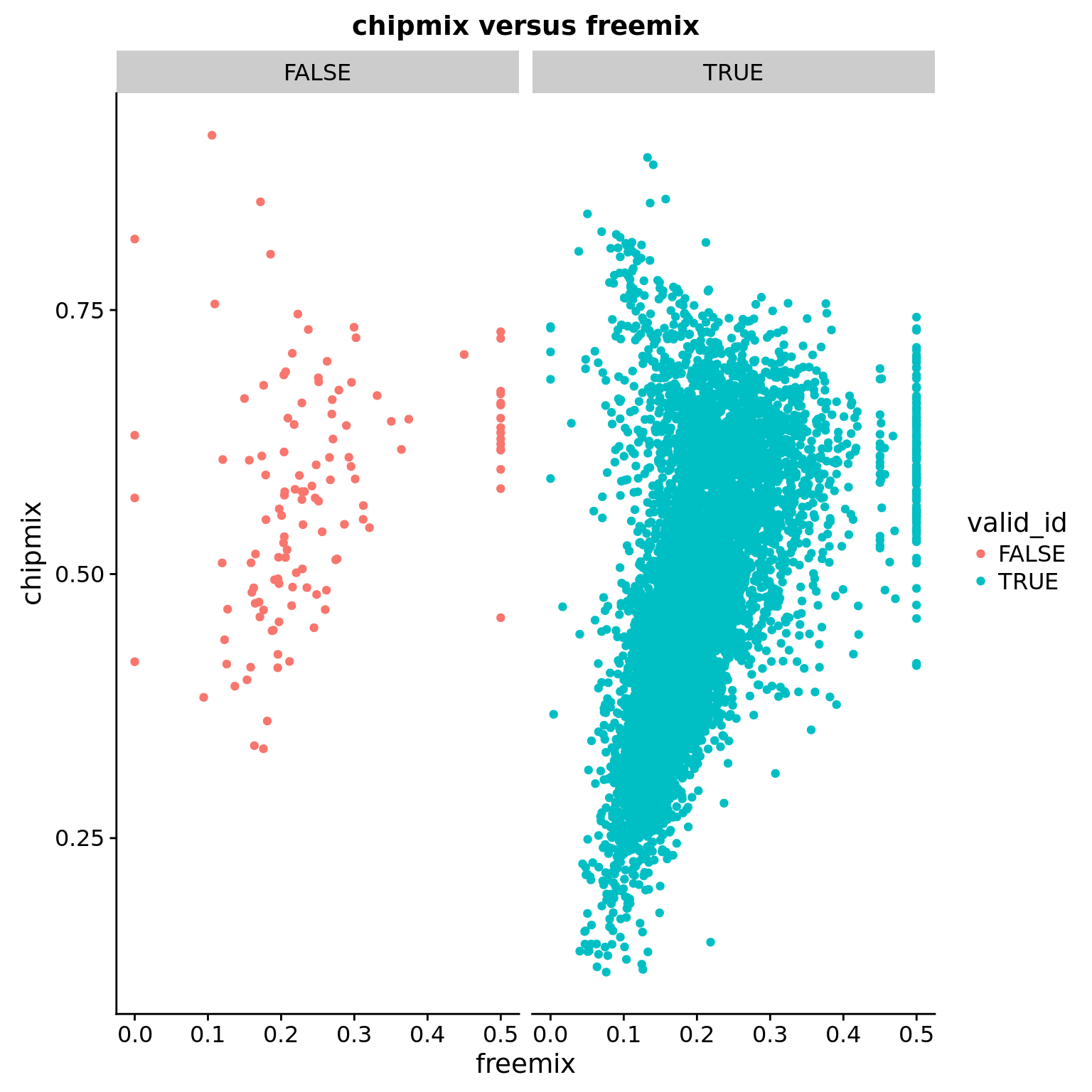
chipmix versus SNPs with overlapping reads
ggplot(anno, aes(x = snps_w_min, y = chipmix, color = valid_id)) +
geom_point() +
facet_wrap(~valid_id) +
labs(title = "chipmix versus SNPs with at least one overlapping read",
x = "SNPs with at least one overlapping read")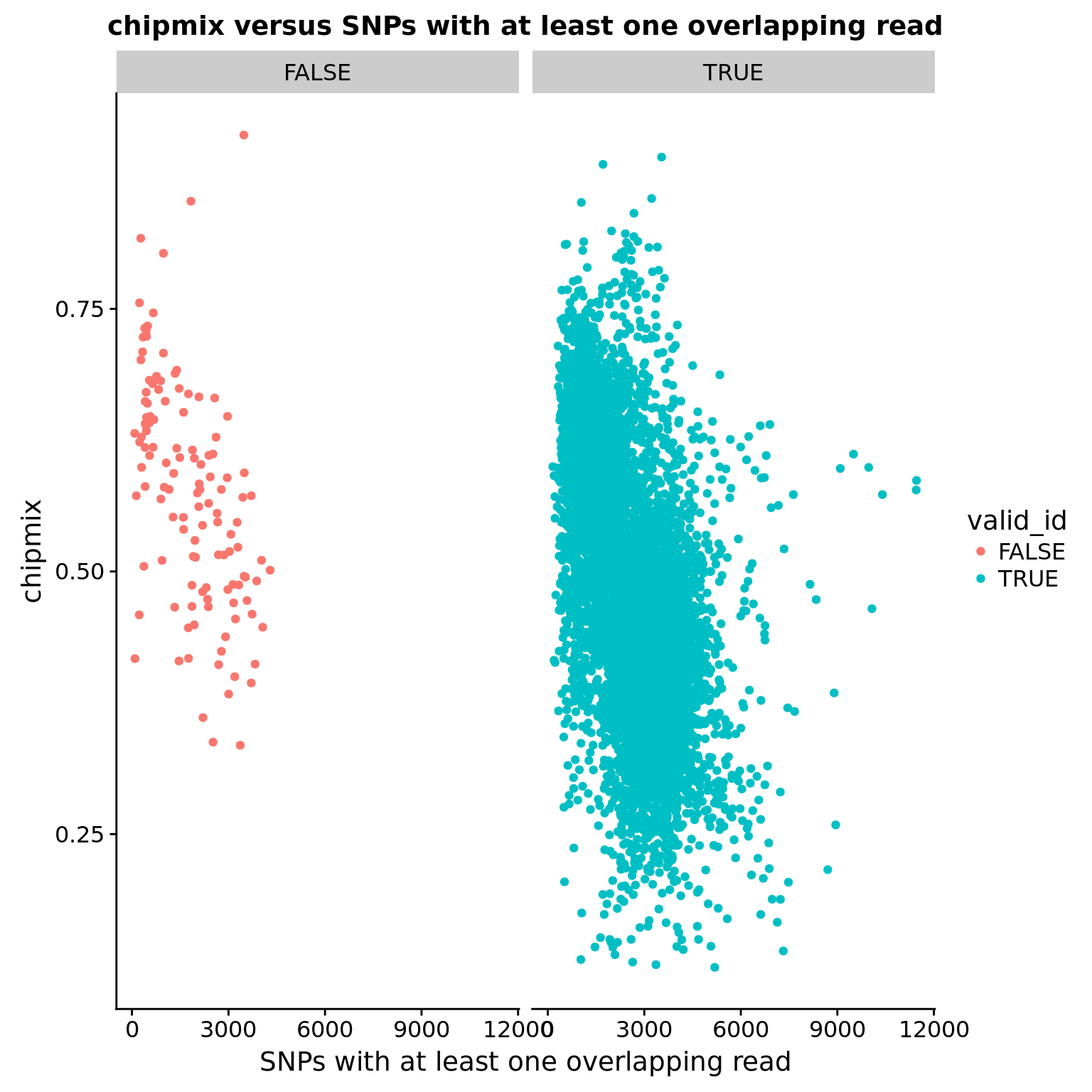
chipmix versus raw number of reads
ggplot(anno, aes(x = raw, y = chipmix, color = valid_id)) +
geom_point() +
facet_wrap(~valid_id) +
labs(title = "chipmix versus number of raw reads",
x = "Number of raw reads")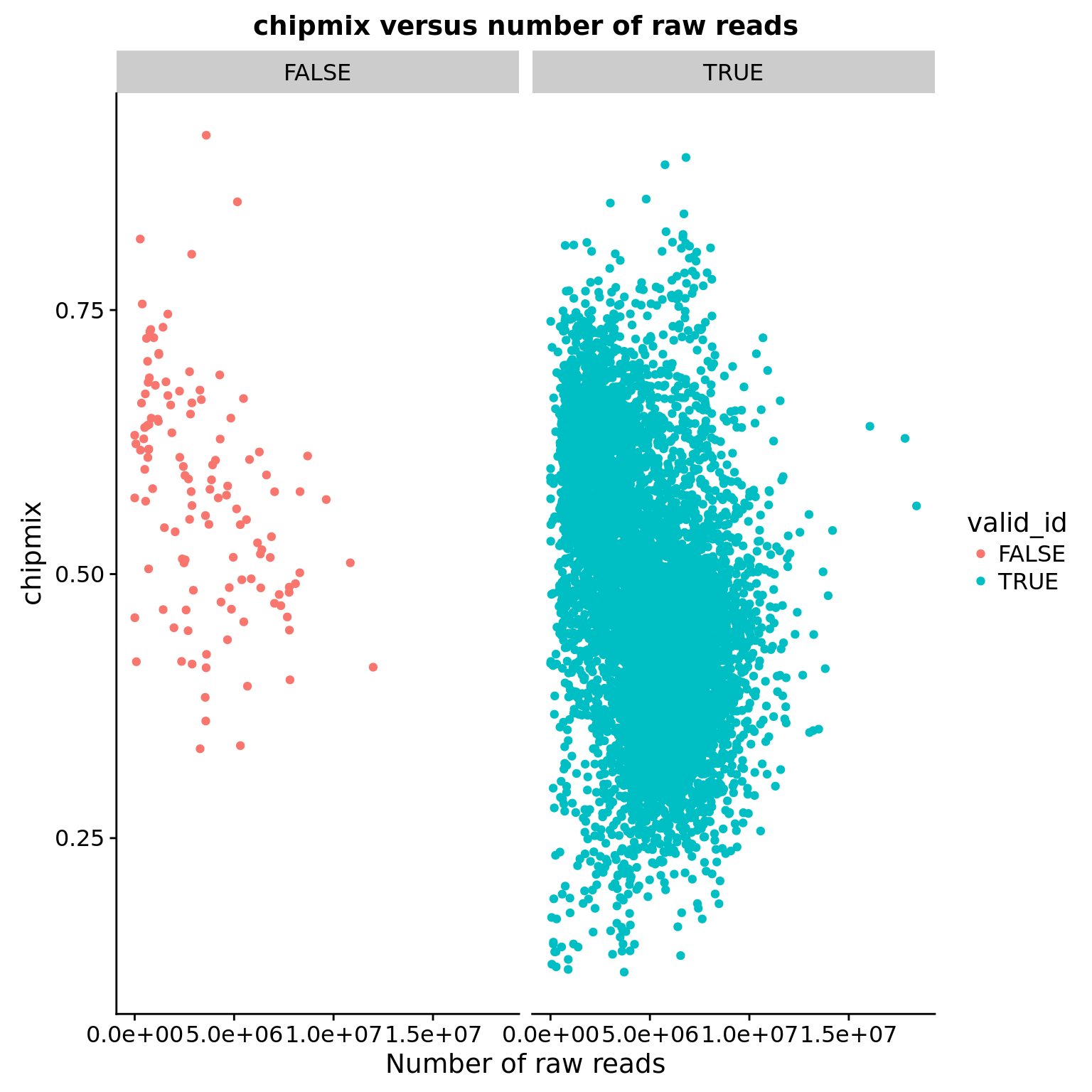
Session information
sessionInfo()R version 3.4.1 (2017-06-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS: /project2/gilad/jdblischak/miniconda3/envs/scqtl/lib/R/lib/libRblas.so
LAPACK: /project2/gilad/jdblischak/miniconda3/envs/scqtl/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel methods stats graphics grDevices utils datasets
[8] base
other attached packages:
[1] bindrcpp_0.2 Biobase_2.38.0 BiocGenerics_0.24.0
[4] tidyr_0.7.1 stringr_1.2.0 dplyr_0.7.4
[7] cowplot_0.9.1 ggplot2_2.2.1
loaded via a namespace (and not attached):
[1] Rcpp_0.12.13 bindr_0.1 knitr_1.20 magrittr_1.5
[5] munsell_0.4.3 colorspace_1.3-2 R6_2.2.0 rlang_0.1.2
[9] highr_0.6 plyr_1.8.4 tools_3.4.1 grid_3.4.1
[13] gtable_0.2.0 git2r_0.19.0 htmltools_0.3.6 assertthat_0.1
[17] yaml_2.1.14 lazyeval_0.2.0 rprojroot_1.2 digest_0.6.12
[21] tibble_1.3.3 purrr_0.2.2 glue_1.1.1 evaluate_0.10.1
[25] rmarkdown_1.8 labeling_0.3 stringi_1.1.2 compiler_3.4.1
[29] scales_0.5.0 backports_1.0.5 pkgconfig_2.0.1 This R Markdown site was created with workflowr