Compares gene annotations between two different cell time types
Joyce Hsiao
Last updated: 2018-07-16
Code version: a6eb777
Goal
Compare cell time estimates derived from algebraic methods versus from trignometric transformation.
Do same for cell times based on FUCCI and cell times based on FUCCI and DAPI.
Getting data
library(Biobase)
df <- readRDS(file="../data/eset-final.rds")
pdata <- pData(df)
fdata <- fData(df)
# select endogeneous genes
counts <- exprs(df)[grep("ENSG", rownames(df)), ]
log2cpm.all <- t(log2(1+(10^6)*(t(counts)/pdata$molecules)))
#macosko <- readRDS("data/cellcycle-genes-previous-studies/rds/macosko-2015.rds")
counts <- counts[,order(pdata$theta)]
log2cpm.all <- log2cpm.all[,order(pdata$theta)]
pdata <- pdata[order(pdata$theta),]
log2cpm.quant <- readRDS("../output/npreg-trendfilter-quantile.Rmd/log2cpm.quant.rds")
pca_double <- prcomp(cbind(pdata$gfp.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust), scale. = T)
pca_triple <- prcomp(cbind(pdata$gfp.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust,
pdata$dapi.median.log10sum.adjust), scale. = T)
library(circular)
theta_triple <- as.numeric(coord2rad(pca_triple$x[,1:2]))
theta_double <- as.numeric(coord2rad(pca_double$x[,1:2]))
names(theta_triple) <- rownames(pdata)
names(theta_double) <- rownames(pdata)Cell times based on FUCCI only
source("../peco/R/intensity2circle.R")
library(conicfit)
par(mfrow=c(1,1))
df <- cbind(pdata$gfp.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust)
ang <- intensity2circle(df, plot.it = TRUE)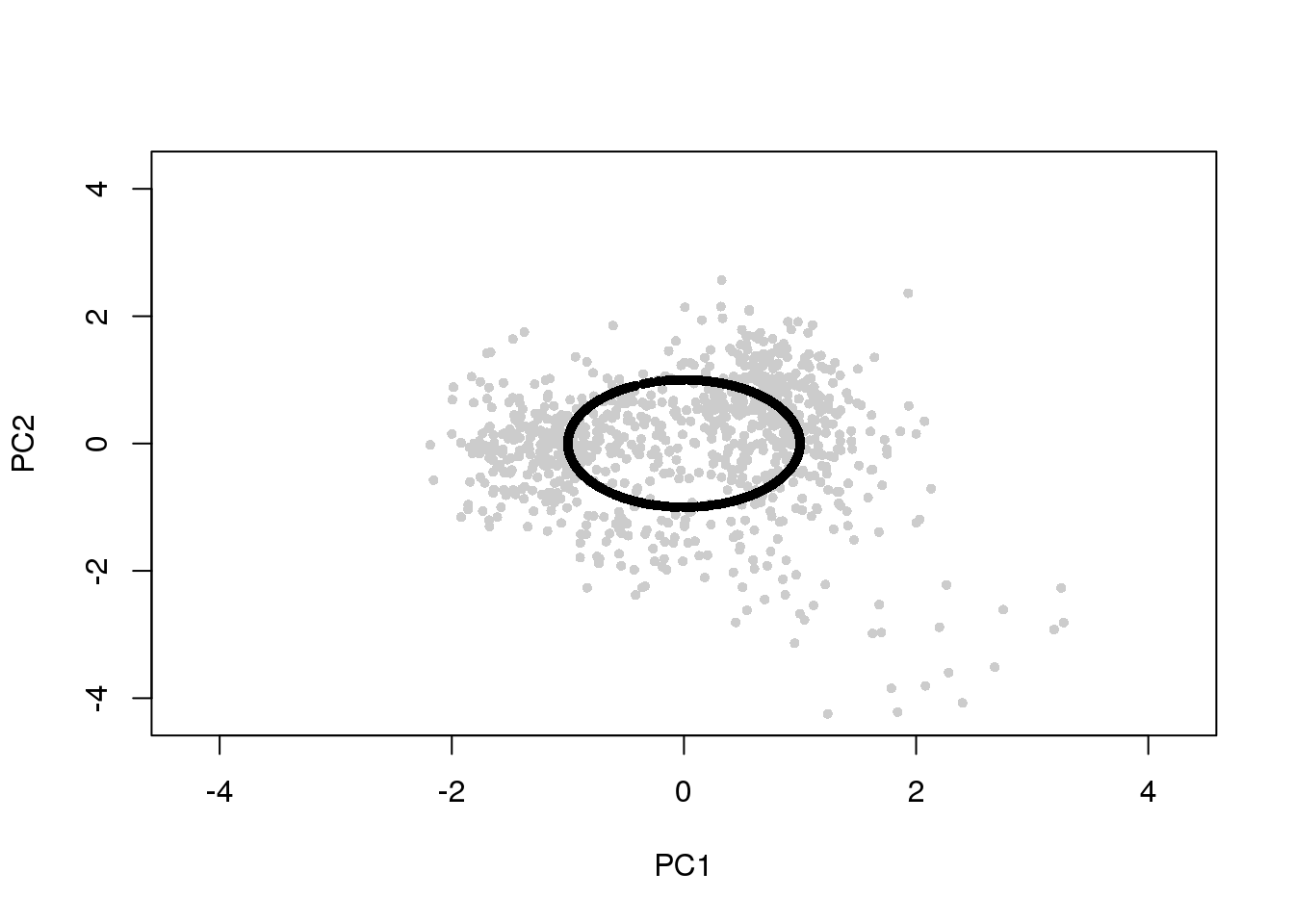
par(mfrow=c(1,1))
source("../peco/R/cycle.corr.R")
ang_shift <- rotation(theta_double, ang)
par(mfrow=c(2,2))
plot(x=theta_double[order(theta_double)],
y=pdata$gfp.median.log10sum.adjust[order(theta_double)], col="forestgreen",
ylab="log intensity", xlab="Cell time estimates",
main = "trignometry", ylim=c(-1, 1))
points(x=theta_double[order(theta_double)],
y=pdata$rfp.median.log10sum.adjust[order(theta_double)], col="red")
points(x=theta_double[order(theta_double)],
y=pdata$dapi.median.log10sum.adjust[order(theta_double)], col="blue")
plot(x=ang_shift[order(ang_shift)],
y=pdata$gfp.median.log10sum.adjust[order(ang_shift)], col="forestgreen",
main = "algebra", ylim=c(-1, 1))
points(x=ang_shift[order(ang_shift)],
y=pdata$rfp.median.log10sum.adjust[order(ang_shift)], col="red")
points(x=ang_shift[order(ang_shift)],
y=pdata$dapi.median.log10sum.adjust[order(ang_shift)], col="blue")
plot(ang_shift, theta_double,
xlab="trigometry-based",
ylab="algebra-based", main="Compar angles"); abline(0,1, col="blue")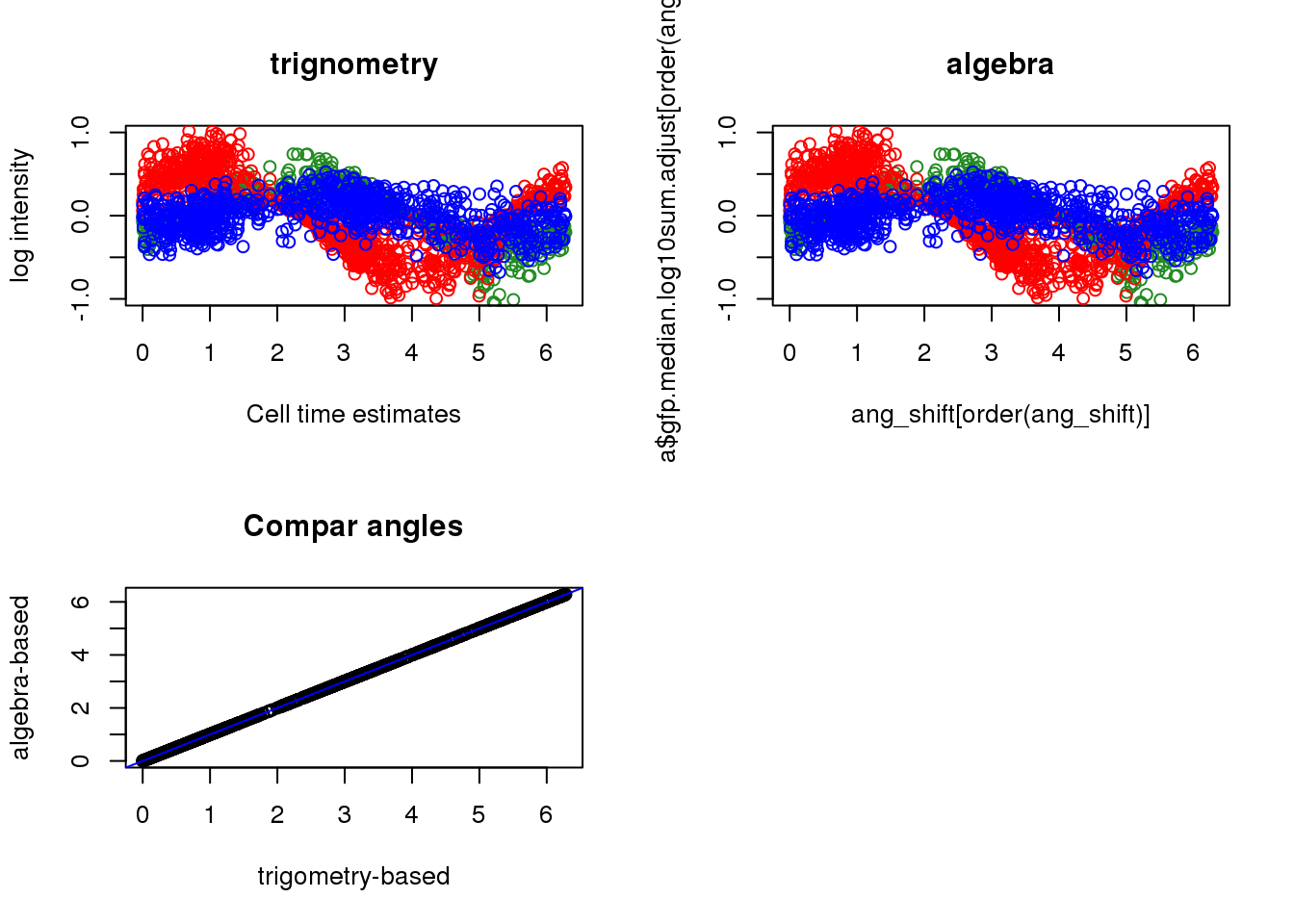
Cell times based on FUCCI and DAPI only
# algebraic methods
source("../peco/R/intensity2circle.R")
library(conicfit)
par(mfrow=c(1,1))
df <- cbind(pdata$gfp.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust,
pdata$dapi.median.log10sum.adjust)
ang <- intensity2circle(df, plot.it = TRUE)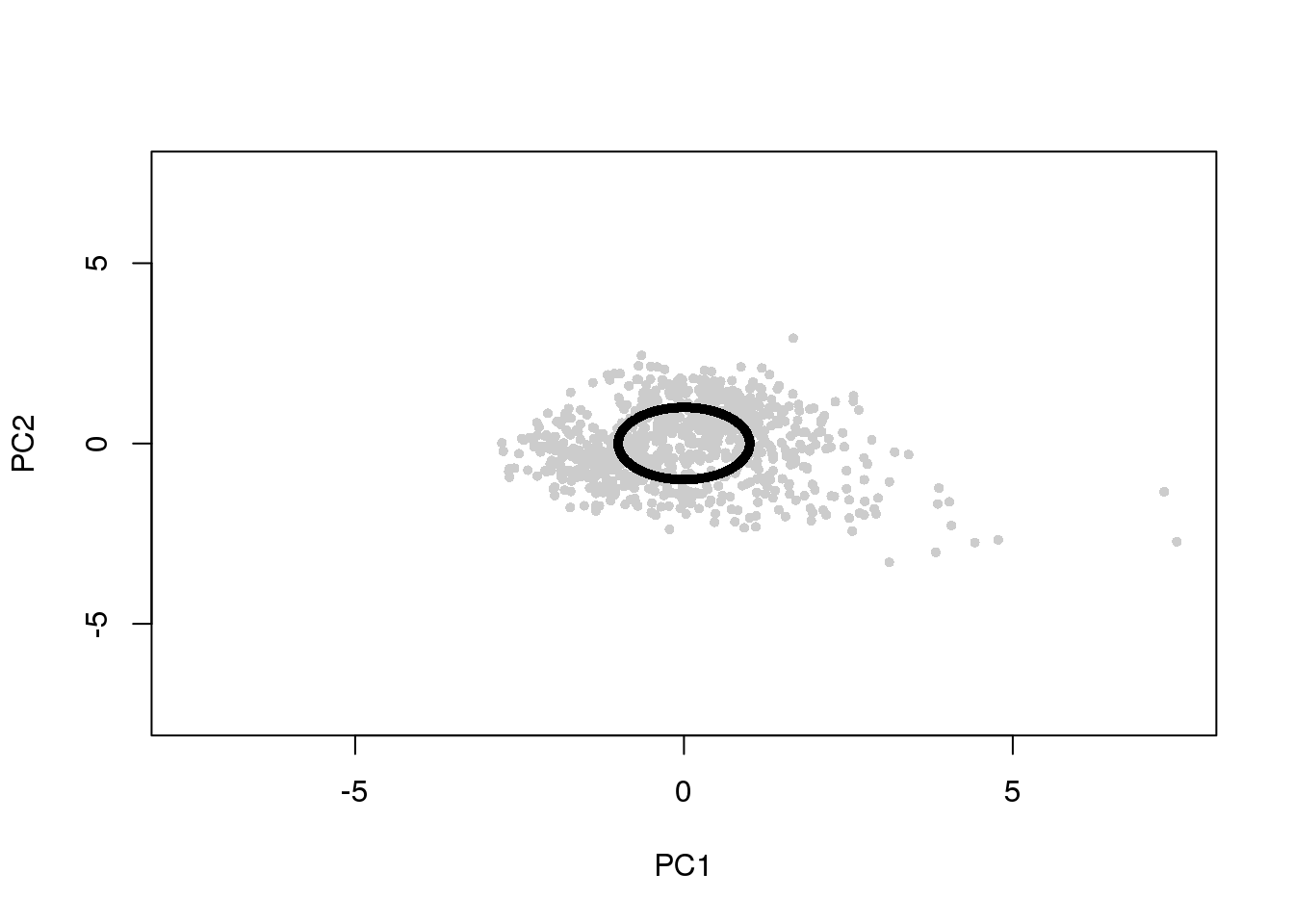
par(mfrow=c(2,2))
source("../peco/R/cycle.corr.R")
ang_shift <- rotation(theta_triple, ang)
ang_shift <- ang_shift%%(2*pi)
plot(x=theta_triple[order(theta_triple)],
y=pdata$gfp.median.log10sum.adjust[order(theta_triple)], col="forestgreen",
ylab="log intensity", xlab="Cell time estimates",
main = "trignometry", ylim=c(-1, 1))
points(x=theta_triple[order(theta_triple)],
y=pdata$rfp.median.log10sum.adjust[order(theta_triple)], col="red")
points(x=theta_triple[order(theta_triple)],
y=pdata$dapi.median.log10sum.adjust[order(theta_triple)], col="blue")
plot(x=ang_shift[order(ang_shift)],
y=pdata$gfp.median.log10sum.adjust[order(ang_shift)], col="forestgreen",
main = "algebra", ylim=c(-1, 1))
points(x=ang_shift[order(ang_shift)],
y=pdata$rfp.median.log10sum.adjust[order(ang_shift)], col="red")
points(x=ang_shift[order(ang_shift)],
y=pdata$dapi.median.log10sum.adjust[order(ang_shift)], col="blue")
plot(ang_shift, theta_triple,
xlab="trigometry-based",
ylab="algebra-based", main="Compar angles"); abline(0,1, col="blue")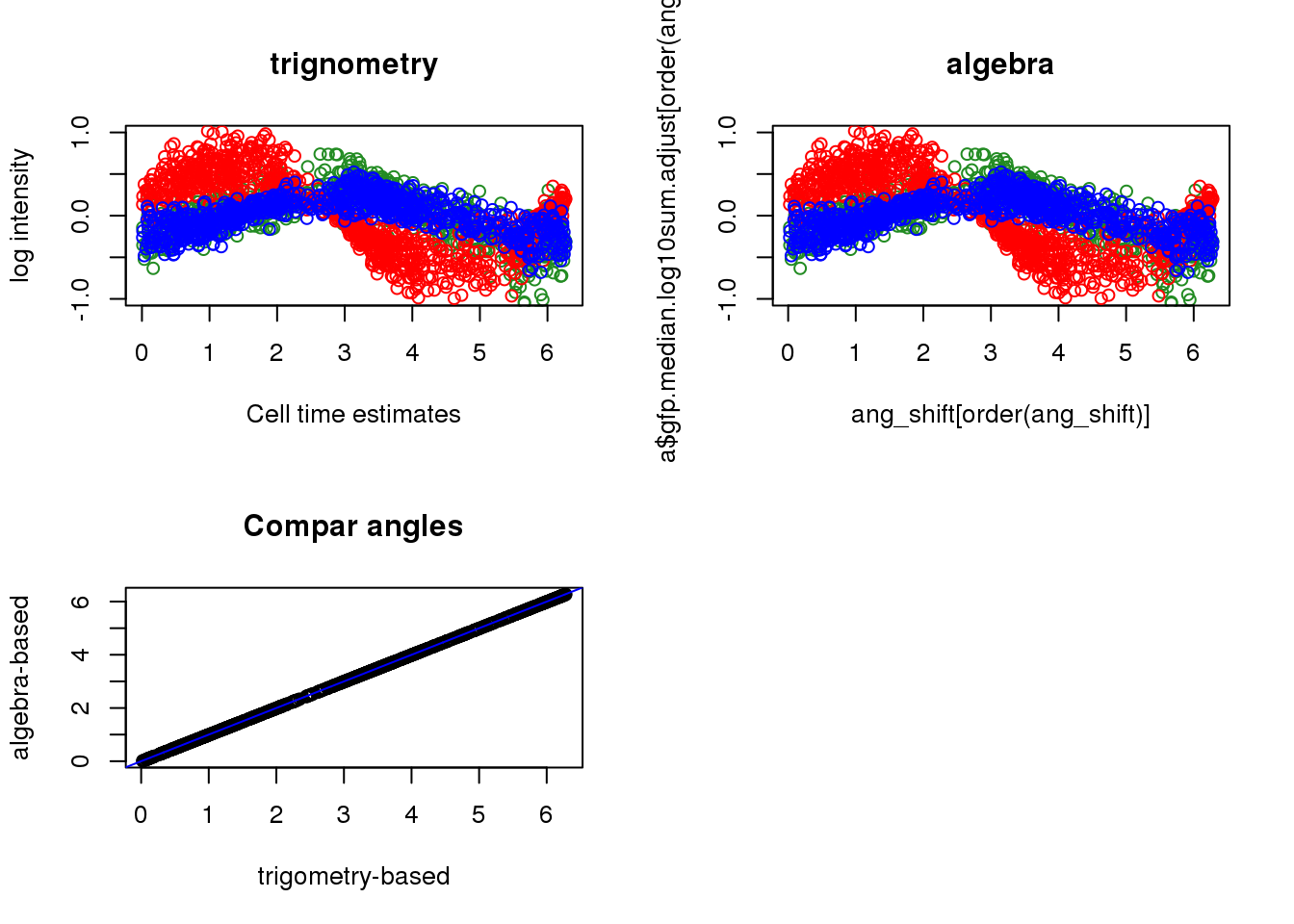
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] conicfit_1.0.4 geigen_2.2 pracma_2.1.1
[4] circular_0.4-93 Biobase_2.38.0 BiocGenerics_0.24.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.17 mvtnorm_1.0-8 digest_0.6.15 rprojroot_1.3-2
[5] backports_1.1.2 git2r_0.21.0 magrittr_1.5 evaluate_0.10.1
[9] stringi_1.1.6 boot_1.3-20 rmarkdown_1.10 tools_3.4.3
[13] stringr_1.2.0 yaml_2.1.16 compiler_3.4.3 htmltools_0.3.6
[17] knitr_1.20 This R Markdown site was created with workflowr