Cyclical trends for expression data of each individual
Joyce Hsiao
Last updated: 2018-09-25
Code version: c6302f1
Summary
Evaluate cyclical trends of expression levels for each gene in each individual
Jobs run on cluster using code in
code/trendfilter-individual.RmdResults saved in
output/trendfilter-individual.RmdThe top 5 cyclical genes are the same across the 6 individuals. When looking at the overlap of the top cyclical genes across individuals, I found that the top 5 cyclical genes are detected in the top 500 cyclial genes across the individuals.
Import results
fits_NA18511 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA18511.rds")
fits_NA18855 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA18855.rds")
fits_NA18870 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA18870.rds")
fits_NA19098 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA19098.rds")
fits_NA19101 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA19101.rds")
fits_NA19160 <- readRDS("../output/trendfilter-individual.Rmd/fits_NA19160.rds")Analysis
pve_NA18511 <- sapply(fits_NA18511, "[[", 2)
pve_NA18855 <- sapply(fits_NA18855, "[[", 2)
pve_NA18870 <- sapply(fits_NA18870, "[[", 2)
pve_NA19098 <- sapply(fits_NA19098, "[[", 2)
pve_NA19101 <- sapply(fits_NA19101, "[[", 2)
pve_NA19160 <- sapply(fits_NA19160, "[[", 2)
pve_NA18511_ord <- pve_NA18511[order(pve_NA18511, decreasing = T)]
pve_NA18855_ord <- pve_NA18855[order(pve_NA18855, decreasing = T)]
pve_NA18870_ord <- pve_NA18870[order(pve_NA18870, decreasing = T)]
pve_NA19098_ord <- pve_NA19098[order(pve_NA19098, decreasing = T)]
pve_NA19101_ord <- pve_NA19101[order(pve_NA19101, decreasing = T)]
pve_NA19160_ord <- pve_NA19160[order(pve_NA19160, decreasing = T)]
names(pve_NA18511_ord)[1:5][1] "ENSG00000148429" "ENSG00000131374" "ENSG00000078114" "ENSG00000096433"
[5] "ENSG00000184349"names(pve_NA18855_ord)[1:5][1] "ENSG00000170312" "ENSG00000175063" "ENSG00000131747" "ENSG00000109654"
[5] "ENSG00000175193"names(pve_NA18870_ord)[1:5][1] "ENSG00000131747" "ENSG00000106853" "ENSG00000165195" "ENSG00000128833"
[5] "ENSG00000177700"names(pve_NA19098_ord)[1:5][1] "ENSG00000170312" "ENSG00000175063" "ENSG00000131747" "ENSG00000117724"
[5] "ENSG00000094804"names(pve_NA19101_ord)[1:5][1] "ENSG00000119812" "ENSG00000111300" "ENSG00000183963" "ENSG00000147596"
[5] "ENSG00000198522"names(pve_NA19160_ord)[1:5][1] "ENSG00000138606" "ENSG00000172175" "ENSG00000100983" "ENSG00000011478"
[5] "ENSG00000077147"library(venn)
intersect(intersect(
intersect(intersect(intersect(names(pve_NA18511_ord)[1:500],
names(pve_NA18855_ord)[1:500]),
names(pve_NA18870_ord)[1:500]),
names(pve_NA19098_ord)[1:500]),
names(pve_NA19101_ord)[1:500]),
names(pve_NA19160_ord)[1:500])[1] "ENSG00000170312" "ENSG00000175063" "ENSG00000102409" "ENSG00000131747"
[5] "ENSG00000154473"checking that trendfilter was done correctly
df_NA19101 <- readRDS("../output/trendfilter-individual.Rmd/df_NA19101.rds")
plot(df_NA19101[which(rownames(df_NA19101)==names(pve_NA19101_ord)[1]),])
points(fits_NA19101[which(names(fits_NA19101) ==names(pve_NA19101_ord)[1])][[1]]$trend.yy,
col="red", pch=16)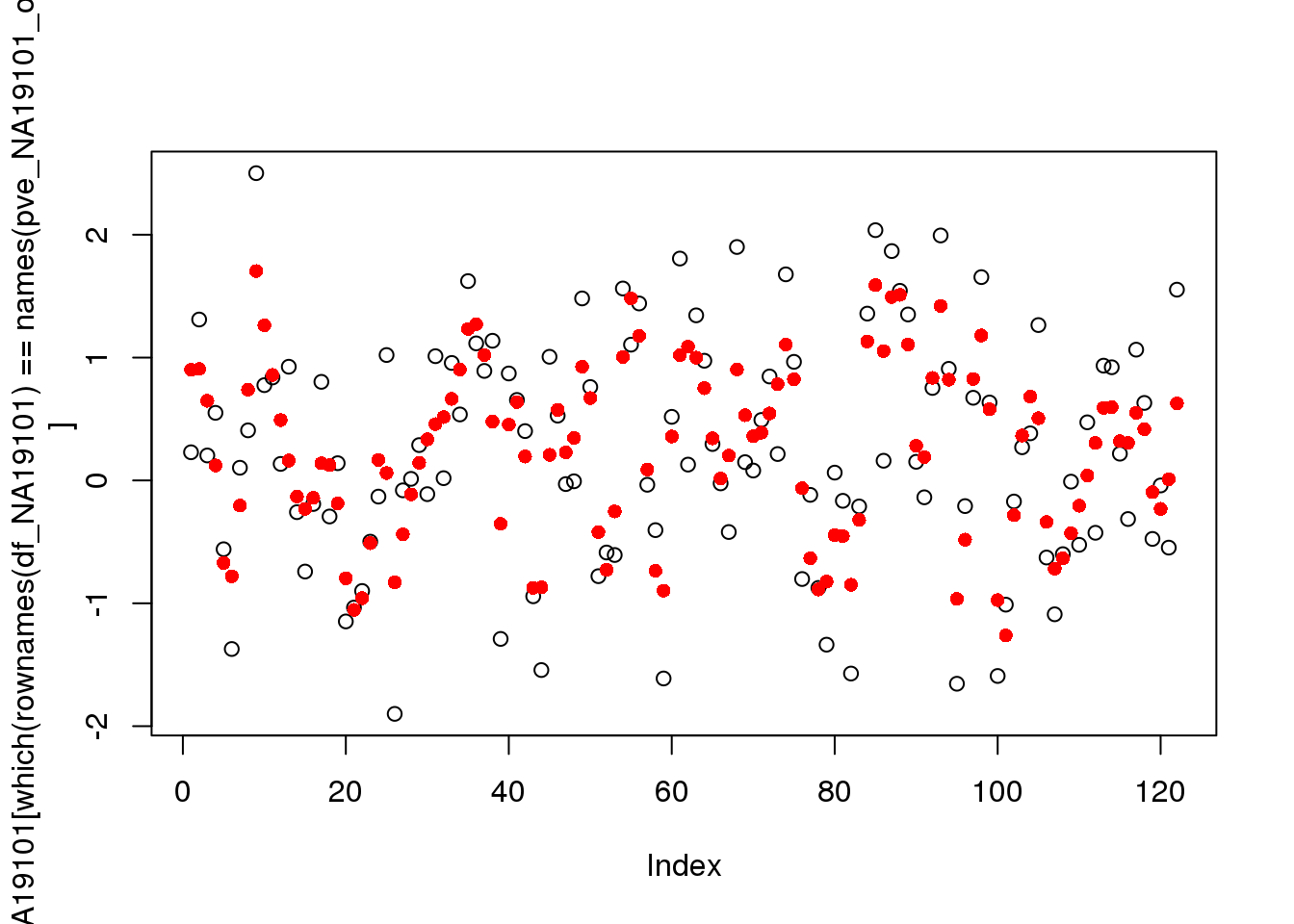
getting PVE of CDK1 from all individuals
pve_NA18511_ord[names(pve_NA18511_ord)=="ENSG00000170312"]ENSG00000170312
0.483625 pve_NA18855_ord[names(pve_NA18855_ord)=="ENSG00000170312"]ENSG00000170312
0.4330661 pve_NA18870_ord[names(pve_NA18870_ord)=="ENSG00000170312"]ENSG00000170312
0.1434487 pve_NA19098_ord[names(pve_NA19098_ord)=="ENSG00000170312"]ENSG00000170312
0.4915681 pve_NA19101_ord[names(pve_NA19101_ord)=="ENSG00000170312"]ENSG00000170312
0.3008477 pve_NA19160_ord[names(pve_NA19160_ord)=="ENSG00000170312"]ENSG00000170312
0.09086933 distribution of cell times in different individuals
eset <- readRDS("../data/eset-final.rds")
library(Biobase)
hist(pData(eset)$theta)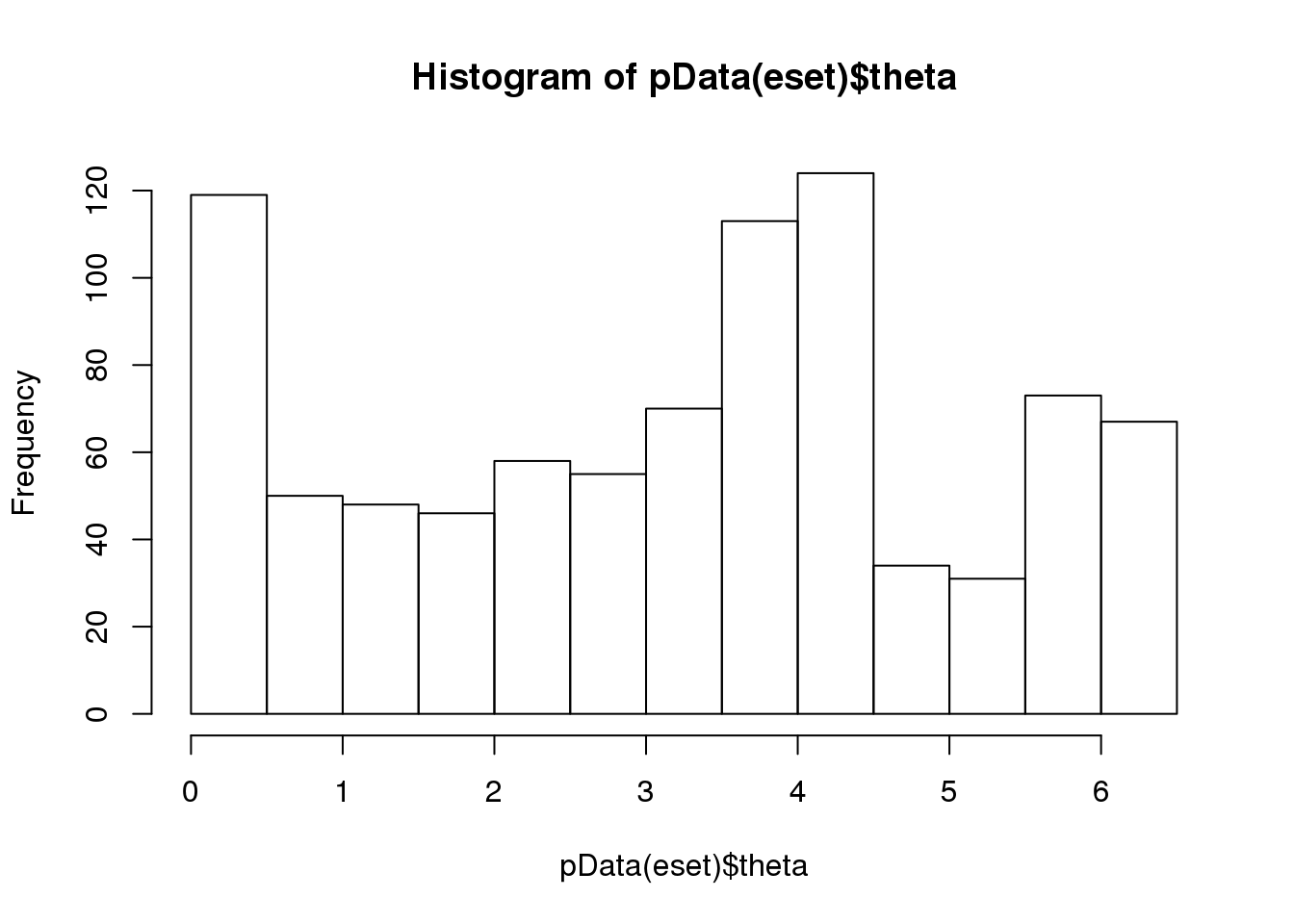
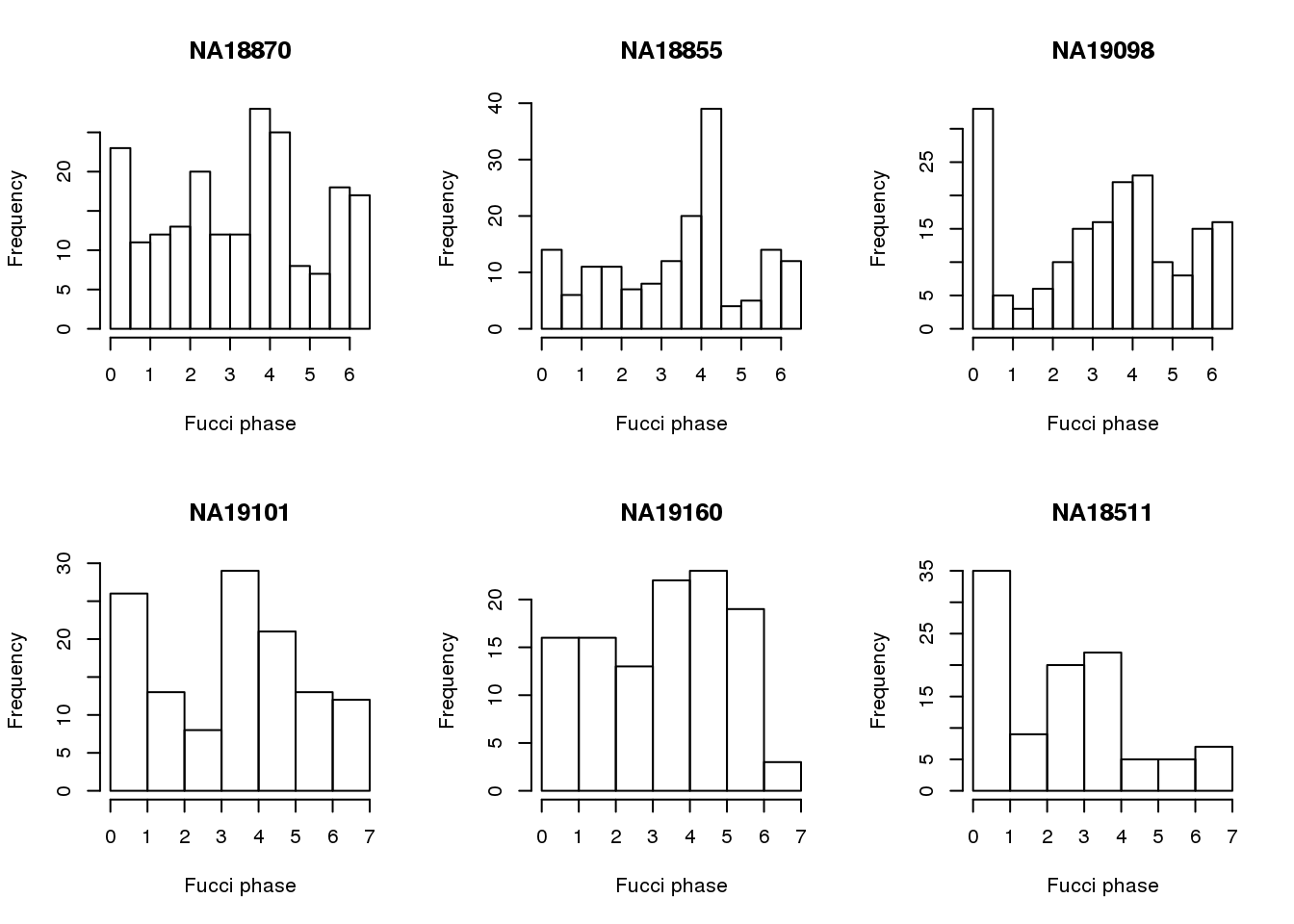
ids <- unique(pData(eset)$chip_id)
par(mfrow=c(2,3))
for (i in 1:6) {
hist(pData(eset)$theta[pData(eset)$chip_id==ids[i]],
main=ids[i],
xlab="Fucci phase")
}
plot(pData(eset)$theta[pData(eset)$chip_id==ids[1]])
points(pData(eset)$theta[pData(eset)$chip_id==ids[2]], col="red")
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] Biobase_2.38.0 BiocGenerics_0.24.0 venn_1.7
loaded via a namespace (and not attached):
[1] Rcpp_0.12.18 digest_0.6.15 rprojroot_1.3-2 backports_1.1.2
[5] git2r_0.21.0 magrittr_1.5 evaluate_0.10.1 stringi_1.2.4
[9] rmarkdown_1.10 tools_3.4.3 stringr_1.3.1 yaml_2.2.0
[13] compiler_3.4.3 htmltools_0.3.6 knitr_1.20 This R Markdown site was created with workflowr